CRYSTAL STRUCTURE OF A ZINC METALLOENDOPROTEASE FROM STREPTOMYCES CAESPITOSUS AT 1.6Å RESOLUTION. G.Kurisu, A.Sugimoto, Y.Kai and S.Harada+, Department of Applied Chemistry, Faculty of Engineering, Osaka University, Suita, Osaka 565, Japan, +Faculty of Pharmaceutical Sciences, University of Tokyo, Hongo, Bunkyo-ku, Tokyo 113, Japan
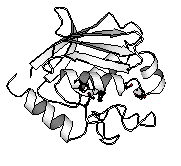
A zinc protease from Streptomyces caespitosus (ScNP), which is specific for peptide bonds on the amino side of aromatic residues, consists of 132 amino acid residues with one disulfide bond. While ScNP has the zinc-binding sequence His83-Glu-Xaa-Xaa-His87, it does not share overall significant similarity to the sequences of other zinc proteases (S. Harada, T. Kinoshita, N. Kasai, S. Tsunasawa and F. Sakiyama, (1995). Eur. J. Biochem. 233, 683-686).We crystallized ScNP and determined its three-dimensional structure at 1.6 Å resolution. The structure analysis was performed by the MIR method. The crystallographic R-factor of the structure refined by XPLOR and PROLSQ was 0.16. ScNP consists of a highly twisted five-stranded [[beta]]-sheet, four [[alpha]]-helices, one catalytically essential zinc ion and one calcium ion as shown in Figure 1.
This structure is topologically similar to those of the catalytic domains of other zinc proteases such as atacin, thermolysin, serratia, snake venom and collagenase despite a lack of sequence homology. The zinc atom of ScNP is tetrahedrally ligated by His83 and His87 in the zinc-binding sequence, Asp93 and a water. ScNP is the first zinc endoproteases in which an aspartate ligates to the zinc, and thus represents a novel organization of zinc ligands.

Figure. 1 Schematic drawing of ScNP1)
1)Kraulis, P.J. J.appl.Crystallogr. 24, 964-950 (1991)