THE DESIGN OF LATTICES FROM DNA BRANCHED JUNCTIONS: PRINCIPLES AND PROBLEMS. N.C. Seeman, J. Qi, X. Li, X. Yang, B. Liu and H. Qiu, Chemistry Dept., New York University, New York, NY 10003, USA.
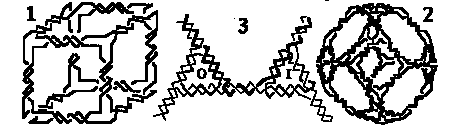
A significant goal of crystallography is 3-D structural control that enables the construction of both individual objects and periodic matter. There are at least three elements necessary for the control of 3-D structure: [1] The predictable specificity of interactions between components; [2] the structural predictability of intermolecular products; and [3] the structural rigidity of the components. The first two of these elements allow for topological control over the products of assembly, in the senses both of the connectivity of the molecular graph and of the linking of plectonemic substructures. The third element, structural rigidity, appears to be needed to fabricate targets that contain symmetry; hence it seems to be particularly important for the construction of periodic networks, whose components exhibit translational symmetry. We are pursuing these ends with synthetic DNA branched junctions. Ligating branched structures generates stick-figures whose edges are duplex DNA, and whose vertices are branch points. Control of topology (elements [1] and [2] above) in this system is strong, and it has allowed us to build DNA molecules whose helix axes have the connectivities of a cube (1 below) and of a truncated octahedron (2 below). To construct lattices, we have sought rigid components, by using DNA triangles whose branches are bulged junctions. By alternating triangles with the bulges on the inside (I) and outside (O) strands, one generates a module with a reporter strand (dark strand in 3 below), whose fate reflects the rigidity of the complex in ligation closure experiments. We have performed such experiments with the components of 3, and find that this system does not satisfy criterion [3]. The search for rigid DNA components continues.

This work has been supported by grants from the NIH and ONR.