 DEFW
DEFW  DEFW
DEFW 
home
software section

DEFW is a simulation program devoted to the simulation of section topographs of dislocations. It must be used only when the incident wave along the entrance surface of the crystal can be considered as a spherical wave (Kato approximation [1]). In the case of plane wave topographs, i.e. when using a good monochromator please use DEF4.
DEFW runs on any modern workstation. It needs less than 1 Mbyte of memory and a moderate time of computation. The result is an image written as non formatted Fortran file. To display it as an image it is usually necessary to convert these data into raw floating point numbers acceptable by most visualisation programs such as Triton or any visualization program written in C language. A program named "convert" is available in the package to convert DEFW images to raw data i.e. to a series of floating point values without any header.
All explanations are given for a Unix machine. However the program may be easily compiled on any PC. A 486 or better processor is recommended.
DEFW is designed to run in batch mode. In most cases the computation needs from a few minutes up to a few tens of minutes without any dialogue, thus it is unnecessary to use an interactive display. All data are edited in a file called topoX.tmp. The name of the file containing the computed image is given in the script file used to start the computation.
A basic script looks like:
# use Korne shell
#
#!/bin/ksh
#
#
# If the image file exists, it must be deleted before starting
# the computation, otherwise the program will stop.
#
#
rm -f image.img
#
# Now we start the program.
# All lines before EOF are data for DEFW. It is limited to the
# name of the image file since all other data are given in a file
# named topoX.tmp, which name cannot be changed
#
defw <<EOF
image.img
EOF
These lines must be written in a file named defw.bat, for instance. Make it executable, using the command: "chmod 700 defw.bat" and submit it as a batch. How to submit a batch depends on your system. Ask your supervisor to know how to do this. You may also run interactively this job by typing the name of the file: "defw.bat" or starting the program and giving the name of the image file.
In this part you will find a brief comment about the data described later in details.
You need:
The elastic stiffnesses are 3 to up 21 independent values - their number depends on the crystallographic symetry of the crystal. You will have to write half the tensor, i.e. you must know which components are null. See for instance [4].
All explanations refer to set of axes described below. You must know:
Note that the Kih are complex numbers and that you must calculate, for each reflection, both the real and imaginary parts. The ratio of the imaginary and real part depend on the choice of the origin in the unit cell. These data are often negative. The program computes the extinction distance and the width at half the maximum of the rocking curve (See the example below) The extinction distance and the width of the rocking curve are a good means to check the consistency of your data.
You must know, from an experimental topograph:
The dimensions of the computed image are computed, as well as the number of pixels along a line or a column, from the knowledge of the thickness of the crystal, the Bragg and the asymetry angle. The width of the image is the width of the beam in the film, for an incidence plane. It corresponds to a line in the image. The height of the image is the vertical dimension. More explanations are given in the section set of axes.
Other data are:
At first look it seems difficult to choose all these data. A simple means to check their consistency is the following: run first the program with the characters: "test" or "TEST" as first characters for the title. The program will write all the output as shown below but it will not run the integration. Thus you will be able to check your data, to verify if the size of the image is reasonnable (most image browsers will not accept more than 512x512pixels), the validity of the diffraction parameters (look at the extinction distance and at the width at half maximum of the rocking curve) and all other data.
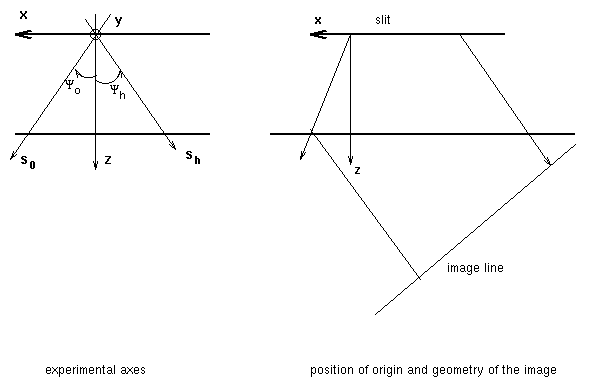
The program uses a set of axes called experimental axes or macroscopic axes. The
macroscopic axes are oriented as follow:
- axis x lies along the entrance surface, oriented from right to left. It is the
projection of the transmitted direction s0 along the surface.
- axis z is the normale to the entrance surface, towards the inside of the crystal.
- axis y is perpendicular to the plane of incidence, in the plane of the surface. The
origin is in the first computed line of the image which means that all lines in the image
correspond to negative y values.
The dislocation orientation may be given either in this set of axes or in the crystallographic axes.
As explained before it is assumed that the film is perpendicular to the difracted beam. Steps are choosen to represent the image in the film, not along the exit surface of the crystal.
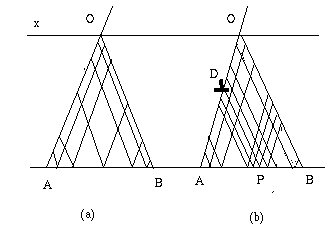
The diffraction equations [3] are a set of partial derivative hyperbolic equations which are integrated using a finite elements method [2]. The integration network, in a plane of incidence, is determined as follow.
The program computes the position of the extinction fringes in a perfect crystal and uses this information to create a network where the local steps vary according to the distance between the fringes [5]. It is minimum along the edges of the Borrmann fan (see fig.2), maximum in the center. A network option, called iopt, allows to choose the minimum and maximum values of the steps. The following table gives their values together with the value of the mean step called Pmean. Pmean is also the reference to choose the resolution in the computed image which is equal to Pmean x kperfo.
| iopt | Pmean | Pmin | Pmax |
|---|---|---|---|
| 1 | 0.8 | 0.2 | 1.6 |
| 2 | 0.4 | 0.1 | 0.8 |
| 3 | 0.8 | 0.1 | 1.6 |
Steps are measured along the exit surface.
Option 1 is usually correct for most computations. However the only method to check the accuracy of the integration of TT equations is to run twice the program using option 1, 2 or 3 and to check if the results are the same. Since the computation time increases tremdously when using the values 2 or 3 this should be done in case of doubt only. Up to now option 1 has been satifying in all cases.
Another option, called, idef, allows to increase (or decrease) the accuracy of the computation. Where the dislocation crosses the transmitted beam (point D in fig.2 b) the phase of the wavefields vary rapidly and it may be useful to increase the precision of the integration by decreasing the integration step in these areas. This is done automatically if idef = 0. The program computes the position of the direct image and switches to the network as drawn in fig.2b in this area. For more explanations see [5]. If idef = 1, the computer ignores the existence of a direct image. Options -1 and -2 remain for compatibility with older versions and should not be used.
This is an example of data file. The numbers on the left, starting at 01, do not belong to the file. They are written to clarify the explanations.
01 Image of dislocation in Silicon
02 5.43 5.43 5.43 lengths of the unit vector of the crystal cell
03 90 90 90 angles for the unit cell (cubic in this example)
04 1 -1 -1 normale to the entrance surface
05 -2 -2 0 diffraction vector
06 -0.3 1 2.11 1 orientation of the dislocation. 1 means macroaxes.
07 0.5 0.5 0 Burgers vector
08 16.6 7.96 7.96 0 0 0
16.6 7.96 0 0 0
16.6 0 0 0
6.4 0 0
6.4 0
6.4 Elastic constants cij (cubic in this example)
09 0.70926 wave length
10 10.667 Bragg angle (positive value)
11 -10.667 Asymetry angle (- theta for symetric reflection)
12 -1.993e-06 -1.57e-08 KIhm dielectric components -h values
13 -1.993e-06 -1.57e-08 KIhp idem h values (real and imaginary parts)
14 300 Height of the image
15 0 Not used. For compatibility with other programs only
16 -32.17 0. 832. Position of core of dislocation in the set of axes
17 0.00142 absorbtion per micron
18 800 Thickness of crystal
19 2 3 0 1 Options: kperfo, imp, idef, iopt
20 0.8 Step of integration for constant step algorithm
(used only if option idef=-2)
Each section refers to the line number in the preceeding part. All input are to be written in free format.
The following example is the contents of to the file defw.log, automatically created when running DEFW, and corresponds to the data shown before.
Program DEFW1. Version June 1993
--------------------------------
Section topograph for dislocation
---------------------------------
Image of dislocation in silicon
Diffraction data
----------------
wave length .709260 Angstroms
Theta .186 Deg.
Psi zero -10.667 Deg.
Mu .001420 per micron
Khi H minus R -1.9930 10(-6), Khi H minus I -.0157 10(-6)
Khi H plus R -1.9930 10(-6), Khi H plus I -.0157 10(-6)
rocking curve half width 2.260 seconds
extinction distance 34.972 microns
Defect Data
-----------
Parameters of the crystal
A = 5.430 angstroms
B = 5.430 angstroms
C = 5.430 angstroms
angles 90.000 90.000 90.000
Burgers vector .500 .500
.000
Dislocation (macro axes) -.300 1.000
2.110
Normale to the surface 1.000 -1.000
-1.000
Diffraction vector -2.000 -2.000
.000
Position of the core (microns) -32.170 .000
832.000
Elastic stiffnesses
16.600
7.960 16.600
7.960 7.960 16.600
.000 .000 .000 6.400
.000 .000 .000 .000 6.400
.000 .000 .000 .000 .000 6.400
Location of direct image 49.078 -270.825
260.559
network of integration
----------------------
Repartition of calculated points along the exit surface
direct image computed
from Y = -257.921 to -283.730 microns
planes number 55 to 76
horizontal step near the defect
till point 17 .20 microns till point 31 .40 microns
till point 51 .80 microns till point 115 1.60 microns
till point 863 .20 microns till point 892 .80 microns
till point 897 .20 microns
Horizontal step far from defect core
till point 17 .20 microns till point 31 .40 microns
till point 51 .80 microns till point 201 1.60 microns
till point 245 .80 microns till point 250 .20 microns
Thickness of simulated crystal 798.49 microns
width of simulated image 295.60 microns
height of image 303.46 microns
vertical calculated step 4.72 microns
horizontal representation step 1.57 microns
vertical representation step 1.57 microns
number of calculated planes 79
number of lines in image 193
Number of points per plane 189
Please note the indication of a version number. It is important to note it in case of error before reporting the problem. The results are presented in different sections as follow:
It is the case in the example where planes 17 up to 31 use this second network.
The name of the file containing the image is not in the data file topoX.tmp. It is part of the script used to run the program or may be input from the keyboard. See the section How to run DEFW.
The image file is written without format:
This file contains some additional informations added by the Fortran subroutines for non formatted write. Thus it cannot be read directly by a visualisation routine written in C language. The program CONVERT is a utility to translate these data in asuitable form.
convert is a utility program to convert the various image formats used in simulation programs. It is self explanatory.
CONVERT allows to:
DEFW and CONVERT sources are available in the directory pub/topo, when logging as anonymous at ftp.lmcp.jussieu.fr/pub/sincris.
Read first 00readme which contains some additional explanations.
To install the program:

Please send your comments and your suggestions to Yves Epelboin, Yves.Epelboin@lmcp.jussieu.fr .
Last update 28/06/98 Y.E.
This service is made available through a grant from CNRS and Ministére de l'Education Nationale
