A.3. 4 cell(F) Cell Constants
Packet Size - 9 words
| SEQ | Description |
| Packet 1 | |
| +1 | a cell dimension in Angstroms |
| +2 | b cell dimension in Angstroms |
| +3 | c cell dimension in Angstroms |
| +4 | cos |
| +5 | cos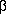 |
| +6 | cos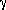 |
| +7 |  in cycles (2 in cycles (2 = 1.0000) = 1.0000) |
| +8 | 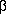 in cycles (2 in cycles (2 = 1.0000) = 1.0000) |
| +9 | 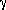 in cycles (2 in cycles (2 = 1.0000) = 1.0000) |
| Packet 2 | |
| +1...+9 | Estimated standard deviations of the quantities of packet 1 |
| Packet 3 | |
| +1...+9 | Reciprocal cell constants, in same order as packet 1 |
| Packet 4 | |
| Transformation matrix from fractional coordinates toorthogonal Angstrom coordinates as given by Rollett (Oxford Computing School,1965), p23. Crystal axes x, y, z; orthogonal axes X, Y, Z are defined with Zalong c, with Y in the b-c plane. | |
| +1 | t11 = a sin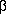 sin sin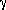 * * |
| +2 | t21 = -a sin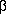 cos cos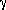 * * |
| +3 | t31 = a cos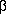 |
| +4 | t12 = 0 |
| +5 | t22 = b sin |
| +6 | t32 = b cos |
| +7 | t13 = 0 t11 t12 t13 x X |
| +8 | t23 = 0 t21 t22 t23 y = Y |
| +9 | t33 = c t31 t32 t33 z Z |
| Packet 5 | |
| +1...+9 | Transformation matrix from Miller indices to orthogonal pseudoMiller indices |
| Packet 6 | Miscellaneous information. |
| +1 | cell volume |
| +2 | e.s.d. of cell volume |
| +3 | last MIR cycle number, initialized to zero |
| +4 | last least squares cycle number, initialized to zero |
| +5 | goodness of fit parameter (chi squared), initialized to 100 |
| Packet 7 | Metric tensor |
| +1 | a2 |
| +2 | a b cos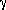 |
| +3 | a c cos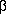 |
| +4 | a b cos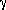 |
| +5 | b2 |
| +6 | b c cos |
| +7 | a c cos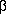 |
| +8 | b c cos |
| +9 | c2 |
| Packet 8 | Inverse metric tensor. |
| +1...+9 | In above order for reciprocal cell parameters |
| Packet 9-10 | |
| Protein data base othogonal to fractional 'scale'transformation as | |
| s11 s12 s13 u1 | |
| s21 s22 s23 u2 | |
| s31 s32 s33 u3, with the last 6 words as 0.0. | |
| Packet 11-12 | |
| Protein data base transformation from fractionalcoordinates to orthogonal (see packets 9-10). | |