3.19. CRYLSQ: Structure factor least-squares refinement
| disper | dispersion factor |
| scale | scale factor |
| extinc | extinction parameters |
| maxhkl | reflection limits |
| xabs | absolute-structure parameter |
| select | select subset of atoms from the bdf |
| dptype | change atomic displacement parameter type |
| group | group or identical molecule refinement |
| atomgr [a] | atoms per group |
| restr [b] | establish conditions for positional parameters |
| constr [b] | constrain one parameter to be a function of otherparameters |
| noref [b] | shut off refinement of specified parameters |
| block | matrix blocking line |
| Notes: a. Must be preceded by a group line b. Order specific lines. restr lines precede constr which precede noref lines. | |
| CRYLSQ | Option | Code | Arg | Def | |
| number of refine cycles | cy | n | 1 | ||
| refinement coefficient | fr | F | |||
| f2 | F2 | ||||
| in | intensity | ||||
| fh | F(heavy atom) | ||||
| derivative matrices | bd | block-diagonal | |||
| fm | full-matrix | ||||
| bl | specified in block lines | ||||
| reflection weight | ws | 1/ 2 2 | |||
| wu | unit weights | ||||
| ww | n | bdf wt with id=190n | 0 | ||
| scale factor(s) | us | refine together | |||
| ss | refine separately | ||||
| dispersion | ad | apply but do not refine | |||
| rd | apply and refine | ||||
| extinction | ax | apply but do not refine | |||
| rx | apply and refine | ||||
| abs-struct. param | aa | apply but do not refine | |||
| ra | apply and refine | ||||
| thermal displacement type | mx | mixed iso and aniso (as in bdf) | |||
| is | isotropic | ||||
| an | anisotropic | ||||
| ov | overall | ||||
| population | pp | yes | |||
| dataset number | ds | n | data set number | bdf or 1 | |
| reflection skip | sk | n | use every n th reflection | ||
| fixed atoms used | ap | apply values from prior run | |||
| ep | apply & store on output bdf | ||||
| type hkl as rcode2 [a] | tl | q | only if Y < q  Y Y | ||
| use in matrix | mr | q | only if w 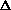 Y < q Y < q | ||
| rcode2 use in matrix | ml | only if Ycal > Yobs | |||
| shift damping factor | fu | q | for bd | 0.8 | |
| for fm | 1.0 | ||||
| non positive-def. U | tr | make pos. def. and continue | |||
| tw | tell user and continue | ||||
| tt | terminate calculation | ||||
| termination criterion | tc | stop if no convergence of R | |||
| list reflections | l1 | last cycle | no | ||
| l2 | every cycle | ||||
| list poor reflections | lr | q | if w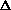 Y >
q Y >
q | ||
| atom lines to pch file | p1 | last cycle | no | ||
| p2 | every cycle | no | |||
| list correl. matrix | m1 | q | if > q after the last cycle | ||
| list deriv. matrices | m3 | after last cycle only | |||
| m4 | after every cycle | ||||
| save correl. matrix | ms | output to file cmx | |||
| old atomic parameters | ao | output old params to the bdf | |||
| Notes: a. Applies for the current run only and does not affect the bdf. | |||||
| disper | 1 | atom-type name | |
| 2 | real part of dispersion scattering factor | ||
| 3 | imaginary part of dispersion scattering factor | ||
| 4-6 | as in 1-3, or use separate disper lines |
| extinc | 1 | extinction correction method | |||
| Zachariasen | zach | ||||
| Becker-Coppens, type I | typ1 | ||||
| Becker-Coppens, type II | typ2 | ||||
| Becker-Coppens, type I and II | gen | ||||
| 2 | extinction type | ||||
| isotropic | is | ||||
| anisotropic (not implemented) | an | ||||
| 3 | mosaic distribution | ||||
| Gaussian distribution | gaus | ||||
| Lorentzian distribution | lore | ||||
| 4 | isotropic extinction parameter(zach or typ1) | (g) | 0 | ||
| 5 | isotropic extinction parameter(typ2 or gen) | (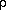 ) ) | 0 | ||
| 6 | mean value of T in mm | Xray | 0.3 | ||
| neutron | 1.5 |
| maxhkl | 1-3 | max |h|, |k|, |l| applied in refinement process | 999 |
| 4 | min value of sin / /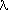 applied applied | 0 | |
| 5 | max value of sin / / applied applied | 10 |
| select | 1- | atom label or atom label string [a] | |
| Notes: a. An atom label string is two atom labels separated by a slash '/'. The first label identifies the first atom , and the second label the last atom in a sequence of atoms. | |||
| dptype | 1 | atom type symbol | |||
| 2 | thermal displacement type | ||||
| overall U | ov | ||||
| isotropic U | is | ||||
| anisotropic Uij | an | ||||
| 3-4 | as in 1, 2, or use separate dptype lines |
| group | 1 | group refinement number of molecule | 0 | ||
| 2-4 | x, y, z fractional coordinate of centre of gravity, if fixed | ||||
| 5 | Euler angle refinement signal | not fixed | |||
| Euler angles invariant | 0 | ||||
| symmetry element is parallel or perp. to a-axis | 1 | ||||
| symmetry element is parallel or perp. to b-axis | 2 | ||||
| symmetry element is parallel or perp. to c-axis | 3 | ||||
| 6 | signal for the refinement of isotropic U | ||||
| group isotropic U | +1 | ||||
| invariant U | 0 | ||||
| individual isotropic U | -1 | ||||
| 7 | signal for population parameter refinement | no constraints | |||
| group population | +1 | ||||
| invariant population | 0 | ||||
| individual population | -1 |
| atomgr | 1 | atom label string [a] | |
| Notes: a. An atom label string is two atom labels separated by a slash '/'. The first label identifies the first atom , and the second label the last atom in a sequence of atoms. | |||
| restr [a] | 1 | restraint class | |||
| bond length | bond | (n=2 atoms) | |||
| bond angle | angle | (n=3 atoms) | |||
| dihedral angle | dihed | (n=4 atoms) | |||
| 2 | atom labels of the n restrained atoms [b] | ||||
| n+2 | ideal values of bond length, angle or dihedral angle | ||||
| n+3 | standard deviation of ideal value | ||||
| Notes: a. restr is an order specific line and cannot be preceded by constr or noref lines. b. These atom labels may each be followed by an optional field indicating a symmetry operation to be applied to the coordinates of the atom. This field has the form (n_mmm), where n is the symop number from STARTX, and mmm is the translation based on the number 555. The parentheses are mandatory. | |||||
| constr [a] | 1 | contraints equation [b] (see CRYLSQ) | |
| Notes: a. constr is an order specific line and cannot be preceded by noref lines. b. Embedded blanks are allowed. The order of the constraints is not arbitrary - reference atoms must occur higher in the atom list than subject atoms. Furthermore, for parameters belonging to the same atom, the reference parameters must occur earlier in the parameter list than the subject parameters. The parameters may be any of the following: SKF, UOV, EXT, DSP, X, Y, Z, U, U11, U22, U33, U12, U13, U23, POP, APP, and NEU. | |||