4.44. PATSEE: Search for molecular fragment
Authors: Ernst Egert and Syd Hall
Contact: Ernst Egert, Institut f�r Organische Chemie, Universit�t Frankfurt, Niederurseler Hang, D-6000 Frankfurt am Main, FRG.
PATSEE searches for a fragment of known geometry in the unit cell using an integrated Patterson and direct methods procedure. This program, which is valid and efficient for all space groups, is based on the standalone program written by Ernst Egert for the SHELX system. The rotation search is applicable to a fragment of any size and allows one torsional degree of freedom. The translation search may locate up to two independent search models of any size (including single atoms), taking into account known atoms at fixed positions, if any. The principles of this method are detailed by Egert & Sheldrick (1985), Acta Cryst. A41, 262-268.
4.44.1. Introduction
The choice of strategy for the solution of a crystal structure at atomic resolution is usually determined by the presence or absence of heavy atoms. Thus it is common practice to solve light-atom structures with direct methods and those containing heavy atoms with Patterson techniques. If thisstrategy fails, it may be advisable to resort to the corresponding alternative method; direct methods may well reveal the positions of heavy atoms, and the Patterson function can be interpreted even for purely light-atom structures, such as those of organic molecules, provided that part of the molecular geometry is known. This so-called Patterson search has been shown by various authors to be a powerful tool for solving difficult crystal structures; its great strength is that it employs chemical information directly, and so can compensate for mediocre precision and resolution of the X-ray data. PATSEE combines the merits of both Patterson and direct methods - in a manner that is generally applicable, efficient, automatic and easy to use - and thus to exploit all the a priori available information in order to solve large problem structures.
4.44.2. Preparation of the Search
Generally, a Patterson search in vector space consists of the following stages: (1) definition of a search model; (2) calculation and storage of the Patterson function; (3) rotation search, and (4) translation search. It is a serial technique, with the last two stages crucially dependent on the accuracy of the preceding ones. Thus the first step is by no means trivial; this is especially true for a procedure such as this where the fragments are taken as rigid and no model refinement is attempted (with the exception of one torsional degree of freedom between rigid groups). Usually a small well-defined search model is more appropriate than a larger one containing several incorrect atoms. The model is defined by atomic coordinates in a given coordinate system; these will normally be either fractional (taken from a related crystal structure) or Cartesian (e.g. from a force-field calculation).
The triplet structure invariant relationships which are required for the translation search (if applied) are calculated prior to the PATSEE run using GENSIN (which gets its E-values from GENEV). The lac1.dat example given below shows a typical input sequence for the GENEV, GENSIN, FOURRFOURR, PATSEE calculations. Note that lac1 is one of the Xtal test decks so that reference to the lac1 listing will be useful in guiding initial PATSEE applications.
The Patterson map is generated by the program FOURR. For almost all purposes, we recommend using E.F as the coefficient ( epat full ); these lead to a sharper map than F2 ( patt full ) but generate fewer ripples than E2-1 ( vect full ).
The region around the origin of the Patterson function is dominated by intra-molecular vectors, which depend on the orientation but not on the position of the fragment. Thus the full six-dimensional search can be split into two three-dimensional searches, a rotation and a translation search (depending on the space group, the latter may be of even lower dimensionality).
The atom fragment information used in the searches is entered in the following way. Atom site coordinates may be entered as fractional or Cartesian according to the option on the preceding FRAG line. Each group of atom sites must be preceded by a FRAG line, or in the case of sites loaded from the bdf, the FRAG line(s) contain the labels of the atom sites to be used in the search. The position of each fragment of the atom sites in the input stream determines how they are used in PATSEE. A fragment that precedes the rotate line will be fixed (i.e. the vectors between these atoms will be neither rotated nor translated but they will be used in the figure of merit calculations). Fragment(s) that follow the rotate line but precede the transl line will be rotated and translated. Fragments that follow the transl line will be translated only. The shift, spin and twist lines are used modified the atom sites in a fragment. If a shift or a spin line will transform or rotate, respectively, the atom sites of the next fragment. The twist line serves a different function -- it enables two parts of a fragment to be rotated about a connecting bond (and searches are applied for each twist settings). The twist line must be positioned in the input stream between the two atom sites which will be twisted with respect to each other.
4.44.3. ROTATE - Rotation Search
The first step in the rotation search is to set up the intra-molecular vector set to be used for the search, i.e. to express the model geometry (which should always be checked thoroughly - see the geom option on the PATSEE line) in the form of discrete vectors with associated weights. Of the N(N-1)/2 intramolecular vectors, the short (e.g. d< p �, see vlim ) and long (e.g. d > q �, see vlim ) values are immediately eliminated. Since the inner sphere around the Patterson origin shows some vector density everywhere, the short vectors provide little angular discrimination and are normally not very useful for determining the orientation of the fragment. However, they may be important for molecules (e.g. those consisting of fused aromatic rings) that are characterized by a few short vectors with high weights. An upper limit for the vector length is also advisable because very long vectors, though quite characteristic of the search model, suffer most from uncertainties in the geometry and could easily miss the corresponding maximum in the Patterson map. Close vectors (see vres ) are replaced by a weighted average vector with the combined weight. In order to save computing time, low-weight vectors may be omitted from the figure of merit assessment (see vfom ).
Any orientation of a rigid fragment relative to a fixed coordinate system can be described by three angles corresponding to successive rotations about properly chosen axes. (There are various definitions of the Eulerian angles. For computational reasons, we prefer successive rotations about the a, b and c axes, in that order.) The asymmetric unit of angular space depends on both the Laue group and the model symmetry. Instead of scanning the respective range of angles by specifying rotation increments, we have chosen to generate random orientations (see nran ). The optimum number of orientations (see ntry ) to be tried depends on the size and the shape of the search fragment, the Laue group and the Patterson grid intervals. PATSEE usually generates 10000-600000 angle triplets, which corresponds to mean rotation increments of about 7�; this is normally sufficient for the coarse location of the maxima.
For each orientation, the correlation between the rotated intramolecular vector set and the Patterson function is measured by a product function (note that this is a different approach to non-Xtal versions of PATSEE). The weight of each vector wi is thus multiplied with the nearest Patterson grid value pi. The rotation figure of merit (Rfom) is ?wi pi for a specified sample (see fraction f of vfom ) of the largest weighted vectors. Note that these vectors are sorted into a test list in the order of descending widi values, where di is the vector length.This is important because a pretest requires that the top t (see vtes ) of the vector list has a Rfom which is positive or the orientation is immediately rejected. The sorting is necessary to avoid the dominance of this test by short heavily weighted vectors. If the final normalized Rfom is < p (see fomt ) the orientation is also rejected.
Before an orientation is placed in the short list of best solutions, it must pass two tests. The 'overlap test' ensure that no close interatomic contacts arise form the application of the lattice translations present and the 'equivalence test' compares the orientation in question with those already stored. Two orientations are regarded as similar when all pairs of equivalent atoms are close to each other; in that case only the better one is kept.
In order to improve the performance of the subsequent translation search, the best solutions are 'refined' by a restricted and finer rotation search. The maximum within each promising region of angular space is found by testing up to n (see nref ) additional random points, which corresponds to a mean rotation increment of less than 2�.
Users may also specify the starting orientation of a fragment (see seta ) and the range or the random rotations (see setr ). This is normally only used, however, if a rotation search is repeated over a target set of orientations.
If the search model has one torsional degree of freedom the rotation searches are repeated for each distinct geometry using the twist option. This specified by a range of possible torsion angles and an appropriate increment. Invocation of twist causes a merged list of best solutions is set up. At the end of the rotation search, a small number of promising orientations are passed over to the translation search. It is our experience that the correct one is usually present among the best two or three for reasonably sized fragments.
4.44.4. TRANSL - Translation Search
In procedures to position a fragment of known geometry in the unit cell, the translation search has usually proved to be less reliable than the rotation search. This is because the 'cross' (i.e. inter-molecular) vectors used to locate a fragment with respect to the origin suffer from errors in both the model geometry and orientation amplified by the symmetry elements; in addition, model vectors with very high weight are less likely than in the rotation search.
The
phases calculated from the coordinates of an oriented model are a continuous
function of the shift vector 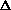 r. When the fragment is moved
through the unit cell keeping its orientation fixed:
r. When the fragment is moved
through the unit cell keeping its orientation fixed:
Fh= Fh�. exp 2  h
h 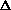 r
r
since all
atomic displacements 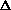 r are the same. So the scattering
contributions from the atoms of the search model have to be summed only once
for each orientation and reflection to yield a structure factor
Fh� for the starting position; subsequently, the
structure factor Fh� for any position is readily
obtained by multiplication with a simple phase factor. For the true structure,
the individual phases of the strongest reflections are linked by various
statistical phase relations; amongst these, the three-phase structure
invariants have proved to be especially useful. The search fragment is usually
incomplete and may also be not very accurate. Nevertheless, if its scattering
power is significant, the triple-phase relations should hold at least
approximately for the correct solution, in the sense that the distribution of
the phase sums is far from being random.
r are the same. So the scattering
contributions from the atoms of the search model have to be summed only once
for each orientation and reflection to yield a structure factor
Fh� for the starting position; subsequently, the
structure factor Fh� for any position is readily
obtained by multiplication with a simple phase factor. For the true structure,
the individual phases of the strongest reflections are linked by various
statistical phase relations; amongst these, the three-phase structure
invariants have proved to be especially useful. The search fragment is usually
incomplete and may also be not very accurate. Nevertheless, if its scattering
power is significant, the triple-phase relations should hold at least
approximately for the correct solution, in the sense that the distribution of
the phase sums is far from being random.
These considerations led us to the development of a novel strategy for a Patterson translation search, which exploits in an integrated fashion the information contained in the sharpened Patterson function, the three-phase structure invariants and allowed intermolecular distances. In short, we have chosen the optimization of a weighted sum of cosine invariants as our refinement procedure, with the Patterson correlation and R indices as additional figures of merit, and the minimum intermolecular distance as a possible rejection criterion. This method is computationally efficient, especially for larger structures, because the refinement is based on phase relations derived from a relatively small number of large E magnitudes (say, >1.8). Only when an acceptable solution has been found by this 'direct search' is it necessary to calculate the time-consuming Patterson correlation.
Since, in order to save computing time, relatively few
phase relations are employed for the refinement, they have to be selected
carefully. Normally only the 40-60 most probable and translation-sensitive
three-phase structure invariants are used for a translation search. It is
advisible to apply a 2 limit to the E values before searching for phase
relations, since high-order reflections may be influenced considerably by
errors in the model. However, if the cut-off is too severe, the accuracy of the
phase-refinement procedure suffers. It seems that a nominal resolution of about
1 � is the best compromise ( smax 0.5 in
GENEV).
limit to the E values before searching for phase
relations, since high-order reflections may be influenced considerably by
errors in the model. However, if the cut-off is too severe, the accuracy of the
phase-refinement procedure suffers. It seems that a nominal resolution of about
1 � is the best compromise ( smax 0.5 in
GENEV).
Then random positions are generated for the rotated search fragment(s); it is our experience that about one translation try per cubic �ngstrom is sufficient in order to have a good chance of locating one search model correctly (see ntry ). Since the number of tries rises as a high power of the number of independent fragments, it is unreasonable to search for more than two fragments simultaneously. However, any number of fixed fragments (obtained from a previous search or a heavy-atom Patterson interpretation, for example) may be added and, in fact, are quite valuable provided their size or scattering power is large enough.
Taking the limited range of the subsequent refinement into account, only those random positions that are fairly close to physically reasonable solutions are worth refining; thus all positions that give rise to short inter-molecular distances (say d < p �) are immediately rejected (see vmin ). The refinement procedure consists of two cycles during which the translation parameters are refined by optimizing t3sum, which measures the triple-phase consistency.
t3sum =  E1E2E3
cos (
E1E2E3
cos ( 1+
1+ 2+
2+ 3)
3)
t3sum is expected to be large and positive for the correct solution. At the end of the second cycle, only positions with t3sum > f (see tprt ) are regarded as possible solutions.
For solutions that have survived these tests, the Tfom value is calculated identically as for Rfom but now for the inter-molecular vectors. A small number of best solutions (according to both t3sum and Tfom) are stored provided that they pass various tests for possible equivalence (allowed origin shift or lattice translation). Although the true position of the search fragment is usually recognizable at this stage, R indices Re1 and Re2 based on E magnitudes have proved very useful in distinguishing further between correct and false solutions.
4.44.5. Combined Solutions
Finally, the solutions are sorted according to a combined figure of merit:
Cfom = (Rfom . Tfom . t3sum 1/2 ) / ( 10 . Re1 . Re2)
For all solutions printed, a Patterson sum function is calculated as a measure of fit/misfit for each individual atom, taking all vectors (intra- and inter-molecular) into account, this enables identification of possible wrong atoms and thus model correction.
4.44.6. Summary
The procedure described differs from other Patterson translation functions in that the oriented model is placed with respect to all symmetry elements of the space group simultaneously. Tests with known structures have indicated that this routine is able to locate very large fragments (of more than 300 atoms), in which case the distance tests sometimes preclude the majority of trial positions, as well as single atoms even when the latter are not very heavy (e.g. phosphorus or sulphur in large organic structures). Above all, the variety of different criteria employed to judge solutions should make this combination of Patterson and direct methods a powerful structure-solving strategy, if chemical information is available. One would expect that a position that is in agreement simultaneously with packing criteria (dmin), the Patterson function (Tfom), triple-phase relations (t3sum) and E values (Re) is probably correct, and our experience shows that this is indeed the case.
4.44.7. File Assignments
Reads symmetry & atom data from the input archive bdf
Writes new atom sites to the output archive bdf
Reads Patterson map from map file
Reads E's and triplets from inv file
Writes new atom sites to file pch
4.44.8. Example
compid lac1 GENEV smax 0.5 list 1.8 :calculate the E values GENSIN :calculate the structure invariants gener 1.8quar noprint 100 1 100 FOURR epat full :calculate the E.F Patterson map PATSEE geom rotate vfom .8 frag 9.946 29.966 11.189 90 90 90 setid site C1 0.43394 0.51778 0.51693 :Diastereoisomer coordinates C2 0.46733 0.54875 0.62187 :Acta Cryst,C39,95 (1983). C3 0.48235 0.52338 0.73773 C4 0.57311 0.48221 0.72824 C5 0.54921 0.45318 0.61956 C6 0.65325 0.41691 0.60175 C7 0.63235 0.38980 0.51082 C8 0.50339 0.39796 0.44173 C9 0.49348 0.44807 0.39978 C10 0.53945 0.48047 0.50109 C11 0.56162 0.45583 0.27891 C12 0.51547 0.42331 0.18395 C13 0.54556 0.37553 0.22118 C14 0.46982 0.36608 0.34010 C15 0.48559 0.31558 0.35594 C16 0.47342 0.29772 0.22711 C17 0.48286 0.33807 0.14279 C18 0.69738 0.36838 0.23249 C19 0.67749 0.50191 0.47513 C20 0.55074 0.326640.02137 O1 0.54488 0.55020 0.82750 N1 0.41764 0.42882 0.62876 N2 0.39747 0.39645 0.53877 setid transl finish |
This is the lac1 test deck. It is the standard test for PATSEE. Use the lac1.dat listing as a guide for other applications of PATSEE.