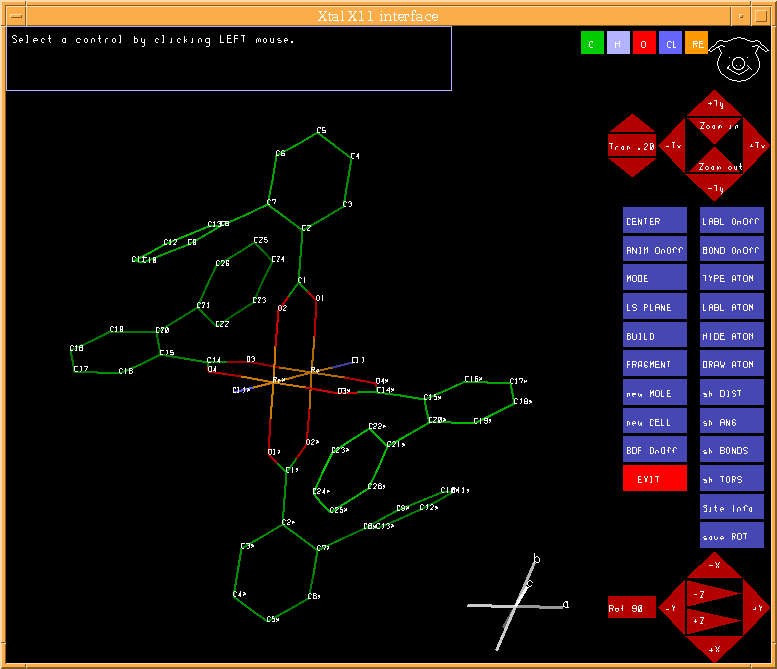
4.46. PIG: Display and manipulate model
Authors: Ursula Bartsch, Syd Hall and Doug du Boulay
Contact: Syd Hall, Crystallography Centre, University of Western Australia, Nedlands 6907, Australia
PIG is a Portable Interactive Graphics routine for displaying, selecting and manipulating atom and peak sites.
4.46.1. Description
PIG extracts atom data from the input archive bdf and/or the peaks data from the pek bdf or the sites from a BUNYIP .bun file. At the conclusion of a PIG session the results of the site manipulations are stored in the lratom: record of the output archive bdf and these may be used directly for subsequent calculations. A list of atom lines is also output to the file add. Note that atom labels are truncated to eight characters in PIG. PIG may be used to display both the molecule and cell. Sites may be rotated, deleted, added, relabelled and retyped. Bond distances, angles and torsion angles may be displayed. The orientation of the the displayed sites is stored on the output archive bdf for use by programs such as ORTEP. The saved orientation matrix is in lrgraf: of the output bdf and will be used by ORTEP as the default plot orientation, unless npig is entered (on the ORTEP line).
The molecular view can be rotated by LEFT-mouse-click-drag-release events. The magnification can be altered by MIDDLE-mouse-click-drag-release events (based on up/down translations) (NOTE that the MIDDLE mouse button is emulated by simultaneous LEFT and RIGHT mouse buttons on two button mice). The molecule can be translated by RIGHT-mouse-click-drag-release events.
PIG can be used as a WYSIWYG (what you see is what you get) interface to POVRAY for raytraced 3D images or ORTEP for traditional 2D thermal ellipsoid plots. Provided that a bdf is written at exit (containing the molecular view orientation), every site visible on screen at the time of exit will be displayed in the subsequent ORTEP/PREVUE. In addition, if labels are switched on, then the ORTEP will be labelled, and if the current view is a unit cell, then ORTEP plots the unit cell outline. Up to 15 ORTEP control lines may be specified in PIG, (prefaced with ORTCTL) and these are substituted for or added to the resultant ORTEP control file commands.
PIG can be used to generate a molecular information file for conversion of XTAL molecular data for use in other programs that do not yet read Crystallographic Information Files (CIFs). Two formats are provided. The Protein Data Bank (.pdb) format, containing space group, cell parameters, symmetry independent sites in cartesian cordinates, anisotropic temperature factors and bond connections. The second format is the Minnesota Supercomputer Centre XMOL .xyz file which contains the cartesian coordinates and atom types of all atoms on screen at PIG exit time (analogous to ORTEP exit mode). The .xyz file maybe useful as a means of generating symmetry related atom sites for use in other software packages such as RASMOL. If the .xyz file is relabelled as a .spf file it can also be used by PLATON (Ton Spek 1980).
More recently any H atoms built in PIG, iether automatically or manually (with the exception of free sites or peak assignments) add constraint linkage information to the archive file. This permits the recalculation of H atoms when invoking the CRILSQ riding model refinement. However, the H atom linkage and labelling is dependent on parent atom coordination. Any attempts to relabel parent atoms or H atoms after building will render the output linkage incorrect. In the event that a riding model refinement is subsequently attempted, it will be necessary for users to iether edit the .add file manually and include it as part of an ADDATM run, or rerun ADDATM in update mode and overwrite existing linkage lines on the archive with their own versions as described in ADDATM.
4.46.2. Interactive Controls
The display is controlled at two levels. (1) Input control lines are used to define the starting parameters for the atom and peak sites to be used, the atom radii, the display colours, the type of display (molecule or cell) and so on. These values are applied prior to displaying the sites. (2) Control is via the interactive cursor and the menu buttons. The user is guided by the instructions provided in the message panel.
4.46.2.1. Rotation Control
Rotation control is provided by 6 buttons in the lower RH corner. The box immediately to the left of this enables the user to switch from course (90o) to fine (adjustable via up down arrows) rotations. These controls are in addition to using the left mouse button.
4.46.2.2. Zoom Control
Zoom and translation control is provided by 6 buttons in the upper RH corner, or alternatively by middle and right mouse button click-drag events.
4.46.2.4. BOND onoff
Selectively hide or display interatomic bond lines. In this mode bonds may be hidden or displayed by clicking on selected labels. The actual change to the display status of the line bond is not effected until the pig icon is clicked (top right hand corner). Note that site markers are NOT hidden with this command, only the connecting bonds (see also the HIDE atoms panel).
4.46.2.5. TYPE atoms
Clicking the cursor on this panel places PIG in type-atoms mode. In this mode the user must first select which atom type by clicking on one of the atom type boxes displayed in the upper right hand area of the display. Then click on each of the sites (to be specific, on each site label) that is to be changed to the selected atom type. The atom type of selected sites will not be changed until the pig icon is clicked. Note that the site labels are not changed on the display, only the colours of the bonds. However, PIG will automatically attach the new atom type code to the atom label when it is output to the pch file and the archive bdf. To change the displayed site label,see the LABL atom command described below.
4.46.2.6. LABL atom
Clicking the cursor on this panel activates the label-atom mode. The user must click on the atom label to be changed and then enter the new label on the keyboard. Duplicate labels will be rejected when exiting from this mode. When labeling is complete click on the pig's nose to exit.
4.46.2.7. HIDE atoms
Clicking the cursor on this panel places PIG in the hide atoms mode. In this mode the user may select any number of sites (by clicking on the appropriate site labels) which are to be removed from the display. The actual removal of site markers, labels and attached bond lines does not take place until the pig icon is clicked.
4.46.2.8. DRAW atoms
Clicking the cursor on this panel will cause all hidden atom sites to be returned to the display. It reverses the HIDE atoms effect.
4.46.2.9. show DISTANCE
Clicking the cursor on this panel places PIG in the show distance mode. A distance will be displayed for two selected sites (by clicking on the appropriate site labels). These sites need not be bonded (this may be used on a 'cell' display as well as a 'molecule' display). This mode is exited by clicking on the PIG icon.
4.46.2.10. show ANGLE
Clicking the cursor on this panel places PIG in the show angle mode. An angle and two connecting distances will be displayed for three selected sites (by clicking on the appropriate site labels). The second selected site is assumed to be the central atom. These sites need not be bonded (this may be used on a 'cell' display as well as a 'molecule' display).This mode is exited by clicking on the PIG icon.
4.46.2.11. show BOND DISTANCES
Clicking the cursor on this panel places PIG in the show bond distances mode. All bond distances connected to a site will be displayed by clicking on the site label.This mode is exited by clicking on the PIG icon.
4.46.2.12. show TORSION ANGLE
Clicking the cursor on this panel places PIG in the show torsion angle mode. The torsion angle and distances will be displayed for the four selected sites (by clicking on the appropriate site labels). These sites need not be bonded (this may be used on a 'cell' display as well as a 'molecule' display).This mode is exited by clicking on the PIG icon.
4.46.2.13. show INFO
Using this mode the user can examine the fractional coordinates of all sites currently displayed, as well as determining the ORTEP/BONDLA symmetry and translation operations required to generate dependent sites, and the peak heights of peaks displayed from PEKPIK data.
4.46.2.14. save ROTATION matrix
Clicking the cursor on this panel causes the currently displayed orientation to be saved in a rotation matrix. On exiting PIG this matrix is stored in the output archive bdf and may be used by ORTEP to produce a plot with the same orientation. Note that doing a LS PLANE calculation will override the save operation.
4.46.2.15. ANIM OnOff
This control activates (by default) or deactivates the animated spin mode. With animation mode activated, using the left mouse button to rotate the molecule via a click-drag-release, imparts an angular momentum to the molecule, proportional to the length and direction of the effective drag displacement vector. To stop a freely spinning molecule simply click and release the mouse button without moving it. Reselect the ANIM control to cancel this mode.
4.46.2.16. CENTRE
The CENTRE control permits re-centring of the molecule, iether about the collective centre of the current molecule, or, about a specific atom site by selecting its label.
4.46.2.17. RG STEREO
Red, green stereo mode. Technically this is an anaglyph. Two images of the current molecule are draw, one of which is rotated by 3 degrees about the molecular centre. By wearing spectacles with one red and one green lens, a 3 dimensional affect may be apparent. Note that if the lens colors are reversed, a freely rotating molecule will appear to rotate backwards, which could be cause for confusion.
4.46.2.18. PEAKS
This control button is only present if (1) the PEAKS command line was invoked from the control file (2) a .pek file exists, or (3) PIG was invoked with the msol multi-solution command. It is a two state button, which switches on or off the display of peaks from a PEKPIK .pek file. However, when peaks are switched ON, a scrollbar facility allows the user to adjust the lower peak eight threshold at which peaks will be displayed. This is useful if there are many insignificant electron density peaks obscuring the larger peaks in which you are interested. The PEAKS button must be reselected to confirm that the current threshold is what the user requires.
4.46.2.19. LS PLANE
Clicking the cursor on this panel places PIG in the least squares plane mode. The user must then select at least three sites (by clicking on the appropriate site labels) to define the least squares plane and then click on the pig icon. This will cause the plane to be calculated, and the atoms sites to be displayed with the plane perpendicular to the z axis. PIG remains in the least squares plane mode and the perpendicular distance of any site from this plane may be displayed by clicking on the site label. This mode is exited by clicking on the pig icon again. Note that the orientation of the plane is automatically stored in the rotation matrix, for subsequent storing on the output archive bdf.
4.46.2.20. FRAGMENT
The FRAGMENT mode has been enhanced over previous versions of PIG. There are now four FRAGMENT choices - select , deselect site connect and site reject . The additional modes were designed primarily as tools for contolling display contents for generating ORTEPS and the .xyz file for export to other programs such as RASMOL.
The select and deselect modes enable all sites within a LEFT mouse button click-drag-release event to be iether exclusively retained or exclusively deleted from the currently displayed molecule. The selected region is highlighted by a rectangular border.
site connect and site reject modes permit the current molecule to be iether extended by adding all bonded sites to a specific atom by selecting its atom label, or by deleting the selected label from the current fragment.
It is important to note that deselecting an atom site or group of atoms using the FRAGMENT modes is quite different from HIDEing an atom in HIDE mode. Using the HIDE button, hidden atoms are irretrievably lost at the next NEW MOLE or EXIT, with potentially disastrous loss of cell contents on the archive file. Whereas, by using the fragment delete modes, the atom sites sites are merely hidden from display. This is why the FRAGMENT mode is preferred as a means of specifying ORTEP plots.
To return to the full molecular display click on the new MOLECULE panel.
4.46.2.21. BUILD
Click on this panel when new sites need to be added to the atom display list. If new H atoms are built, on exit they are assigned Uiso values corresponding to 1.5 or 1.25 times the Uiso value of their bonded C/N/O parent (if present), dependent on whether it is a methyl group or otherwise, respectively. Because of this it is more generally useful to have refined the vibrational motion of the heavier atoms before building H atoms.
Five additional panels will be displayed in the message area. Clicking on one of these panels places PIG into a particular 'build atom site' mode. For each mode follow closely the instructions displayed in the message area. The modes are:
Linear Requests a new site which is linearly aligned with two other sites in the structure. First select an atom type for the new site. Then click on the two sites aligned with the new site; the furtherest site first and then the site to which the new site will be bonded. The new site will automatically be assigned a site label composed of the label of the bonded site with the character 'a' appended.
Trigonal Requests a new site which is trigonally aligned with three other sites in the structure. First select an atom type for the new site. Then click on the three sites which form an apex to which the new site will be attached; the apex site is clicked second in the sequence. The new site will automatically be assigned a site label composed of the label of the bonded site with the character 'a' appended.
Tetrahedral Requests one, two or three new sites which are tetrahedrally bonded with neighbouring sites in the structure. First select the atom type for the new site(s). If one or two sites are sought, click on the three sites which form an apex to which the new site(s) will be attached; the apex site is clicked second in the sequence. If three tetrahedrally related sites are sought (e.g. methyl hydrogens) click on the two sites which form the fourth member of the tetrahedral bonding; the furtherest site first and then the site to which the new sites will be bonded The display will be then aligned so that the bond of the two clicked sites will point directly at the viewer and be surrounded by a circle. The position of the three new sites around this bond is selected by clicking the position of the first at a point on this circle. All new site(s) will automatically be assigned a site label composed of the label of the bonded site with the characters 'a', 'b' or 'c' appended.
Free site Requests a new site which is not geometrically related, but is bonded, to another site in the structure. First select an atom type for the new site. Click on the site to which the new site will be bonded. Then click on the screen to determine the orientation (relative x and y coordinate; z will be set at the value of the bonded site). Note that the display usually needs to be carefully oriented before this mode is used. The new site will automatically be assigned a site label composed of the label of the bonded site with the character 'a' appended.
Auto H atom In this mode PIG searches every independent C atom in its atom array and examines its connectivity, based on current radii values. If the C atom is underbonded its bondlengths and angles are assessed for linear, trigonal and tetrahedral H atom bonding. Where there are no ambiguities, these H atoms are automatically built and labelled according to their parent C atoms label. No H atoms are built on parent O atom or N atoms because the number (for N) and orientation (for O) of such H atoms are in general not well defined. In general, bond angles below 116 degrees are treated as tetrahedral, and C-C bond lengths below 1.46 Angstroms are considered double bonds.
4.46.2.22. RADII
Use the RADII button to set the atomic radii for bonds to be calculated in future. Formerly the radii button was subsumed with the new MOLE command, but it is now accessible from the main menu to assist with fragment extension. In RADII mode selecting an atom type box will display the current radius, and reselecting that atom type enables a new value to entered via the keyboard (closed with a return carriage).
4.46.2.23. HIDE TYPE
This control is used to temporarily hide specific atom types from view, such as hydrogen atoms in cluttered diagrams. When invoked, selecting an atom-type panel immediately switches the display state of all sites of that type on or off. Select PIG to cancel this mode. Atom types hidden in this manner are not lost and may be redrawn later by repeating the same procedure.
4.46.2.24. new MOLECULE
Clicking this panel causes the currently displayed sites (not the hidden sites) to be included in the calculation of a new connected molecule and its display. Hidden atoms (from Hide Atom) will be removed from the site list and will not be available for output to the archive bdf. Conditions for the connected molecule search are set by the PRESET line and by the three panel buttons that appear in the message area. These buttons control the following:
SYM-EXT on/off - a toggle switch to include/exclude atom sites related by symmetry which are within the bonding distance. This switch should be on if a molecule contains a symmetry element but may need to switched off when peaks are active.
Clusterng - A three position switch to enable/disable the reclustering of sites, or revert to the input site list (losing all changes) when the molecule is redrawn. The cluster mechanism attempts to optimally connect all input sites to the first atom site in the input list. For very large molecules clustering is time consuming. If a molecule contains more than 500 atoms, clustering is automatically switched off.
Cluster Packing- A four position switch governing the treatment of multiple disconnected clusters. The first mode (Pack Off) switches off any extra packing treatment. The three additional packing modes provide variations on the optimal packing of secondary clusters about the primary aggregate molecule. Pack close attempts to arrange clusters in order to minimize distance between the centres of gravity of all clusters. Pack centre attempts to minimize the distances between the largest cluster center of gravity and other clusters Pack contact attempts to find the shortest contact between clusters - to accentuate hydrogen bonding for example.
Clicking the New Molecule panel a second time will start the display process using the current display parameters.
4.46.2.25. new CELL
Clicking the cursor on this panel causes the currently displayed sites (not the hidden sites) to be included in the calculation of a new cell display. The mcut option allows for all molecules to be cut at the cell boundary of the first unit cell, but otherwise all sites within +/-1 cell along all three axes are calculated and added to the display view. Hidden atoms (using HIDE ATOM) will be removed from the site list and will not be available for output to the archive bdf, or the add file. Note that the current display limit is 10000 atoms. If nine unit cells result in more sites than the current limit, then PIG will terminate with an error message.
4.46.2.26. POV IMG
This mode provides the user with limited control over the creation of compid.pov files containing a three dimensional description of the current display for rendering using the freely available Persistence of Vision Ray Tracer(TM) (Pov-Ray). XTAL is in no way associated with POV-Ray(TM) and makes no warranties as to that programs reliability, usefulness, or suitability for any purpose whatsoever. Consequently no support is offered or will be given towards installation of such external software. That being said, POV-Ray is a very powerful visualization tool and its use is recommended. In addition (under unix), if the environment variable POVEXE is set to the location of the POV-Ray(TM) executable, POV-Ray(TM) will be automatically invoked as a background process and the current scene rendered, on screen and also to the file compid.ppm.
In POV IMG mode the user can adjust several options by selecting the apropriate controls in the PIG message window. When satisfied, the user should reselect POV IMG to create the .pov file and render it - or choose PIG to cancel the mode. In addition, if labels are switched on in PIG then the output image will be labelled, and if the unit cell is being drawn, then it too will be rendered in the POV-Ray(TM) image.
CPK model This control sets the desired molecular representation to either close packed overlapping spheres (CPK), Ball and stick model, Rods or thermal ellipsoids.
400x300 This is the smallest of five preset image dimensions. Rendering time increases significantly with image dimension.
No Octants This is intended for ellpsoid plots where the principle axes may not be obvious.
Prob 50% This sets the default probability level for rendering thermal ellipsoid plots.
Users should note that the ASCII text compid.pov file generated by XTAL includes important header information from the xtalpov file located in the directory specified by the XTALHOME environment variable. However, if a user has an alternate xtalpov file in the current working directory, it will be used by default. In this way image renderings can be customised by the user, setting different backgrounds, adjusting perspective, scaling and atom colors. Also the xtal.pov files generated by XTAL are sufficiently well commented that they can be edited by users to delete selective atoms, their labels or bonds, and redisplayed by manually running POV-Ray(TM) from the command line.
4.46.2.27. BDF on/off
Clicking on this panel toggles the output mode of the archive bdf between on and off. The default mode is on. If the mode is on, the displayed atom sites (including the retained peak sites and labels), and the orientation matrix of the displayed sites, will be stored on the archive file by the program ADDATM. Independently of the bdf mode, a file .add is output containing all displayed atom sites and the display orientation.
4.46.2.28. EXIT
Clicking on this panel will commence the exiting process from PIG. First the display will be redrawn in the currently stored orientation. If the user wishes to sort the output atom list based on atomic number and labels, there are three possible label SORT modes. The user can choose one mode and continue, or ignore and continue by reselecting the EXIT button. At this point the user has the option to proceed with an ORTEP/PREVUE process based on the current screen display, (assuming that a new bdf will be written) or to complete the exiting process by clicking on the EXIT button again. An accidental exit> can be canceled by selecting the PIG icon.
4.46.2.29. NEXT/PREV
When PIG is automatically invoked after CRISP, as a structural solution viewer, (run as PIG msol from the command line) two additional control buttons are invoked between the message panel and the atom-type buttons. These buttons permit the stepping forwards and backwards through structural solutions loaded from the .sol file produced by CRISP.
4.46.3. Sort modes
The three atomlist sort modes all sort sites according to atomic number, but they differ in their label sorting algorithms. The method to choose for sorting labels is largely dictated by the structure and the site labelling scheme adopted. It is inevitable that the atom sites written to the .add file will require additional manual ordering for publication purposes, but the three modes provided sould provide a helpful starting point. The different schemes are best illustrated by way of example.
sort left sorts with the leftmost label character most significant
atom F2 .28990 .19669 1.00840 .035 atom F3 .28330 .25008 .79120 .035 atom F32 .35080 .42862 .47520 .035 atom F33 .48430 .37205 .31320 .035 atom F35 1.12030 .28096 .45740 .035 atom F36 1.01950 .33188 .62620 .035 atom F5 1.02790 .17466 .67020 .035 atom F6 1.08261 .12219 .87690 .035 |
sort right sorts with the rightmost label character most significant
atom F2 .28990 .19669 1.00840 .035 atom F32 .35080 .42862 .47520 .035 atom F3 .28330 .25008 .79120 .035 atom F33 .48430 .37205 .31320 .035 atom F5 1.02790 .17466 .67020 .035 atom F35 1.12030 .28096 .45740 .035 atom F6 1.08261 .12219 .87690 .035 atom F36 1.01950 .33188 .62620 .035 |
sort numeric sorts labels into numeric order
atom F2 .28990 .19669 1.00840 .035 1.00 atom F3 .28330 .25008 .79120 .035 1.00 atom F5 1.02790 .17466 .67020 .035 1.00 atom F6 1.08261 .12219 .87690 .035 1.00 atom F32 .35080 .42862 .47520 .035 1.00 atom F33 .48430 .37205 .31320 .035 1.00 atom F35 1.12030 .28096 .45740 .035 1.00 atom F36 1.01950 .33188 .62620 .035 1.00 |
4.46.4. File Assignments
Reads atom data from the input archive bdf
Optionally reads the BUNYIP file bun if the buny
option entered. Optionally reads peak data from bdf pek
Optionally reads the CRISP file sol if the msol option is entered
Writes final connected atom sites to line file add and thereby to the output archive bdf
Writes final cartesian coordinates to the line file pdb
Writes display cartesian coordinates to the line file xyz
Writes POV-Ray description file pov
4.46.5. Examples
Enter atom data from archive bdf
Enter atom and peak sites from file pek. Set the peak site radius to 1.0 Angstrom, and the atom type radii for Zn and Cl atoms to 1.7 and 1.2 Angstroms, respectively.
PIG ort : invoke PIG with a view to ORTEP generation radii Zr 1.7 Cl 1.2 : set bonding radii ortctl exec ialab nhla : override automatic ORTEP line option ortctl vsc 1 1 2 2 5 1.5 1.6 : Add an extra vsc instruction ortctl genins axes 2 : override automatic genins line ortctl elipse 6 3.37 : override elipse line finish |
Use PIG as a dedicated ORTEP plot specifier with additional explicit ORTEP controls to fine tune the output. Subsequently run PREVUE to edit label positions and potentially invoke PLOTX from PREVUE to generate Postscript output.