Elementary X-ray diffraction for biologists
Jenny P. Glusker
1. General teaching
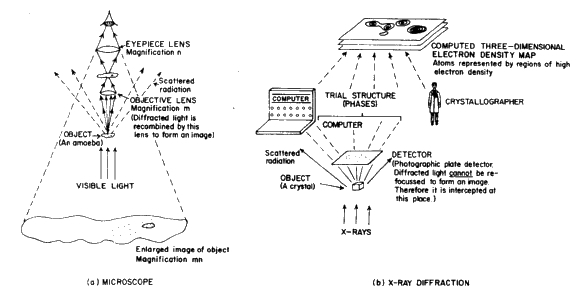
The results of X-ray diffraction studies of both small and large molecules of biological interest have greatly enhanced our understanding of many biochemical processes such as enzyme mechanisms, nucleic acid flexibility and virus assembly. Since biologists are primarily interested in biology, if a physical method is described to them, it is important that it be carefully explained how the results can be of use in understanding some aspect of biology. Since most biology students are very familiar with the microscope, some of the analogies described below are very useful (e.g. Fig. 1). Some ideas for a short 10 minute exposition and for a short course are given below. It is best to have lots of visual demonstrations.
a. 10 minute talk
Have on hand a fine sieve or piece of gauze, a point source of light, a diffraction photograph of DNA fibres and a model of DNA. The talk begins with a demonstration of diffraction. This is done by having the students view the point source of light through the sieve or gauze. Then it is shown how the diffraction pattern is formed as a result of the regularity of the grid and constructive or destructive interference of scattered waves by this. If the mesh size is then varied, the reciprocal relationship between the mesh and the spacing of the diffraction pattern can be shown. This can also be demonstrated by showing diffraction photos of sodium chloride and a protein on the same scale.
The audience is then told that the whole experiment may be scaled down so that the sieve is replaced by a crystal (with spacings of the order of 10-8 cm) and that the visible light is replaced by X-rays (with wavelengths of the order of 10-8 cm). During this explanation it will be necessary to give some description of the regularity of a crystal as a result of the build-up of unit cells.
The next stage is to display a DNA model and Rosalind Franklin's DNA photo (mentioning the books The Double Helix by Watson and The Eighth Day of Creation by Judson). All regularities of structure that could cause a diffraction pattern are pointed out. There is a 3.5 Å spacing between the DNA bases along the helix axis, and this accounts for the large spot at the top and bottom of the X-ray photograph of DNA (Fig. 2). The parallel lines of backbone in the helix have a regularity that explain the cross in the middle of the photograph. Since the distances in DNA are larger for this regularity, the spots on the X-ray photograph are closer together.
It is hoped that the audience will, on leaving, know what a diffraction effect is, and what DNA looks like.
b. Course of 4 hours (usually with lots of solicited student interruptions)
1st hour. Diffraction is discussed, as above. If white light is used to produce the diffraction pattern, some splitting of the spots into red and blue is seen, and this effect can be used to explain the wavelength effect on the diffraction angle. It is then explained how the diffraction pattern is measured (detectors, cameras, geometry, etc.) and the types of diffraction patterns obtained (usually precession or oscillation photographs or diffractometer measurements). This can also be done with a model, such as a collimator stuck into a styrofoam brick (X-ray tube), a goniometer head with a dummy crystal mounted on a wire, and precession photograph (preferably Polaroid, mounted on a wooden stand). The heights of each should be appropriately adjusted so that an imaginary X-ray beam passes through the collimator, hits the crystal, and then falls on the centre of the film (marked to indicate a beam catch for the direct beam). The physics of diffraction and the concepts of path differences and orders of diffraction are then explained. Make sure at the end of this that the audience knows what a diffraction data set is.
2nd hour. Then the audience is told how structures are derived from the diffraction data set. First, the reason why some 'reflections' are intense is explained in terms of structure (for example, the DNA photograph and model could be used). Then the Patterson map is described in detail. The best simple description is given by Judson in The Eighth Day of Creation. Imagine a party and that at a given instant everyone's shoes were stuck to the floor. If each person then shook hands with every other person how would he turn, how far must he stretch and in total how may interactions would there be? This introduces the concept of vectors very simply. It is helpful then to analyse the Patterson map of a simple structure and one containing a heavy atom. Also it is shown how the orientations of groups of known structure may be deduced by comparing (and reorienting if necessary), a calculated and observed Patterson map.
Then the use of direct methods is described for the simple (centrosymmetric) case of the 1 0 0 and the 2 0 0 reflection (both intense, hence 2 0 0 has a phase of 0![]() ) (see Glusker and Trueblood, Crystal Structure Analysis: A Primer ). This explanation can be expanded to more general reflections if the audience is sufficiently interested. Then the use of isomorphous replacement is described. This can be illustrated elegantly by optical diffractions (with a laser light) of repeating patterns reduced to a suitably small size (for example, Sung Hou Kim uses a drawing of a duck, and of various eggs as the different heavy atoms). The intensity variation on isomorphous replacement (an egg vs. no egg) can then be seen. (See Atlas of Optical Transforms and Pamphlet No. 1 which describe experiments on optical diffraction in detail.)
) (see Glusker and Trueblood, Crystal Structure Analysis: A Primer ). This explanation can be expanded to more general reflections if the audience is sufficiently interested. Then the use of isomorphous replacement is described. This can be illustrated elegantly by optical diffractions (with a laser light) of repeating patterns reduced to a suitably small size (for example, Sung Hou Kim uses a drawing of a duck, and of various eggs as the different heavy atoms). The intensity variation on isomorphous replacement (an egg vs. no egg) can then be seen. (See Atlas of Optical Transforms and Pamphlet No. 1 which describe experiments on optical diffraction in detail.)
3rd hour. This time is most effectively spent by repeating the second hour's lesson. Usually most students have understood by the second time around.
4th hour. The methods of refinement are described, i.e. difference maps and particularly the method of least squares (using a simple linear equation to illustrate this method graphically). The estimation of precision (not to be confused with accuracy) of the result is then described.
The types of results and the information in crystal structure publications, particularly protein structure papers, are then described. One useful way to help the student understand descriptions of protein folding is to have him thread beads of different colours (one colour for ![]() -helix, another for pleated sheet, etc.) on a string and then fold the string of beads as described in the article.
-helix, another for pleated sheet, etc.) on a string and then fold the string of beads as described in the article.
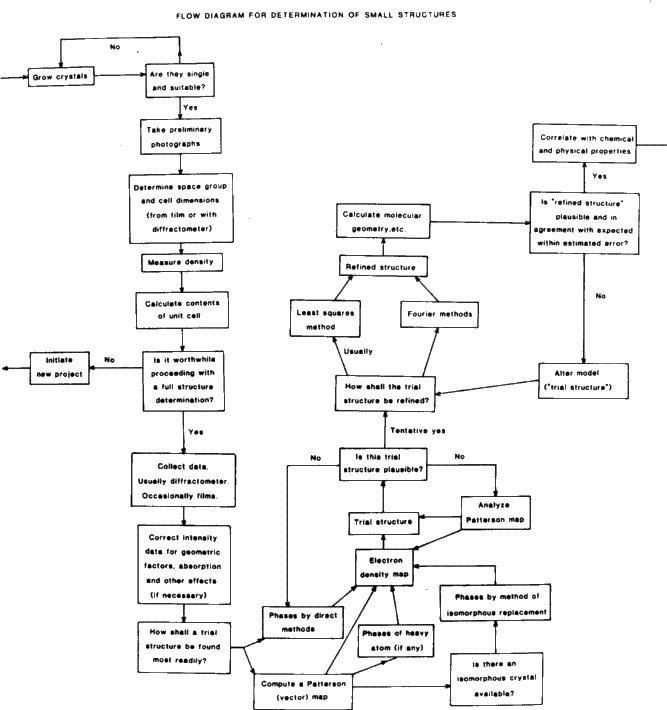
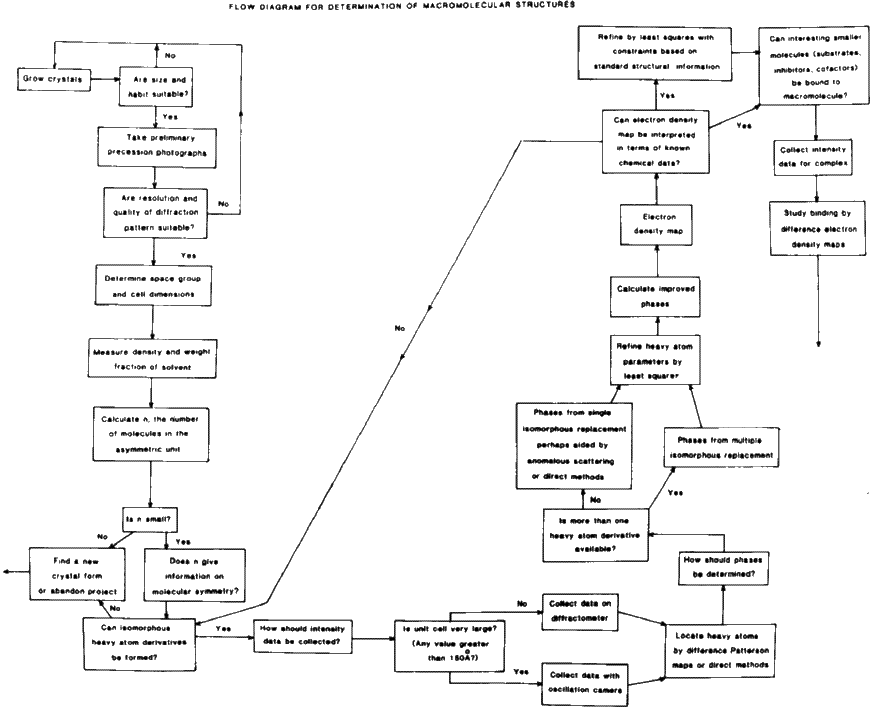
Some flow charts for structure determination are shown in Figs. 3 and 4.
2. Some textbooks
a. Lehninger, Albert L., Biochemistry, 2nd edition, New York, Worth Publishers, Inc. (1975).
b. Stryer, Lubert, Biochemistry, San Francisco, W. H. Freeman and Co. (1975); second edition (1981).
c. Schulz, G. E. and Schirmer, R. H., Principles of Protein Structure, New York, Heidelberg, Berlin, Springer-Verlag (1979).
d. Cantor, C. R. and Schimmel, P. R., Biophysical Chemistry. I. The Conformations of Biological Macromolecules. II. Techniques for the Study of Biological Structure and Function. III. The Behaviour of Biological Macromolecules, San Francisco, W. H. Freeman (1980).
e. Cold Spring Harbor Symposia on Quantitative Biology. Volume XXXVI. Structure and Function of Proteins at the Three-dimensional Level, New York, Cold Spring Harbor Laboratory (1972).
f. Dickerson, R. and Geis, I., The Structure and Action of Proteins, Menlo Park, Benjamin/Cummings (1969). (A revised edition is in preparation entitled Proteins: Structure, Function and Evolution. )
g. Fersht, A., Enzyme Structure and Mechanism, Reading and San Francisco, W. H. Freeman (1977).
h. Alworth, W. L., Stereochemistry and its Application in Biology, New York, London, Sydney, Toronto, Wiley-Interscience (1972).
i. Watson, J. D., Molecular Biology of the Gene, 3rd edition, Menlo Park, California, W. A. Benjamin Inc. (1976).
j. Blundell, T. L. and Johnson, L. N., Protein Crystallography, London, New York, San Francisco, Academic Press (1976).
k. Glusker, J. P. and Trueblood, K. N., Crystal Structure Analysis: A Primer, New York, Oxford University Press (1972) (second edition in press).
l. Stout, G. H. and Jensen, L. H., X-ray Structure Determination. A Practical Guide, New York, The Macmillan Company (1968).
m. Dunitz, J. D., X-ray Analysis and the Structure of Organic Molecules, Ithaca and London, Cornell University Press (1979).
n. Vainshtein, B. K., Modern Crystallography. I. Symmetry of Crystals, Methods of Structural Crystallography, Berlin, Heidelberg, New York, Springer-Verlag (1981).
o. Glusker, J. P., Structural Crystallography in Chemistry and Biology, Stroudsburg, Pennsylvania, Hutchinson Ross Publishing Company (1981).
p. Also see the movie 'Life and the Structure of Hemoglobin'.
3. Molecular dimensions
It is important that some basic principles of conformation be taught to students. They are added here for completeness.
Accuracy is a measure of the deviation of an observed quantity from its correct value. Since we never know the truly correct value we don't know how accurate our data are.
Precision is a measure of the experimental uncertainty in a measured quantity, an indication of its reproducibility. This is what we measure and is what we indicate in an e.s.d.
Precision of Measurements. It is important to stress that a value quoted as 1.395(4) Å means 1.395 ![]() 0.0004 Å where
0.0004 Å where ![]() means an estimated standard deviation (e.s.d.) (derived from the inverted least squares matrix used in refining the structure). If the distribution of errors is normal, then there is a 99% chance that a given measurement will differ by less than 2.7 times the e.s.d. from the mean. A bond length of 1.542(7) Å (1.542 Å with an e.s.d. of 0.007 Å) is probably not significantly different from one measured at 1.527(7) Å.
means an estimated standard deviation (e.s.d.) (derived from the inverted least squares matrix used in refining the structure). If the distribution of errors is normal, then there is a 99% chance that a given measurement will differ by less than 2.7 times the e.s.d. from the mean. A bond length of 1.542(7) Å (1.542 Å with an e.s.d. of 0.007 Å) is probably not significantly different from one measured at 1.527(7) Å.
a. Bond lengths and angles
Bond lengths in Å. C--C 1.54, C=C 1.34, C![]() C 1.20, C--C aromatic 1.39, C--O 1.43, C=O 1.23, carboxylic acid 1.30, 1.20, carboxylate 1.26, C--N 1.47, C--N in heterocycles 1.35, C--H 1.09, C--S 1.81, C=S 1.71. Tetrahedral angles sp3 109.5. Trigonal angles sp2 120.0
C 1.20, C--C aromatic 1.39, C--O 1.43, C=O 1.23, carboxylic acid 1.30, 1.20, carboxylate 1.26, C--N 1.47, C--N in heterocycles 1.35, C--H 1.09, C--S 1.81, C=S 1.71. Tetrahedral angles sp3 109.5. Trigonal angles sp2 120.0![]() . Linear bonds sp 180.0
. Linear bonds sp 180.0![]() . Octahedral coordination 90
. Octahedral coordination 90![]() .
.
b. Torsion angles
The torsion angle (or angle of twist) about the bond B--C in a series of bonded atoms A--B--C--D is defined as the angle of rotation needed to make the projection of the line B--A coincide with the projection of the line C--D, when viewed along the B--C direction. The positive sense is clockwise.
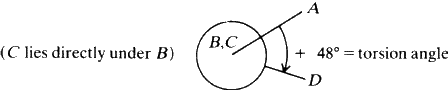
In general a chain of --CH2-- groups will have a staggered conformation so that torsion angles are 180![]() for C--C--C--C and 60
for C--C--C--C and 60![]() for C--C--C--H or H--C--C--H.
for C--C--C--H or H--C--C--H.
c. Molecular packing
Hydrogen bonds X--H![]() Y are 2.38 to 3.20 Å with the proviso that the H
Y are 2.38 to 3.20 Å with the proviso that the H![]() Y distance should generally be 2.2 Å or less. The X--H
Y distance should generally be 2.2 Å or less. The X--H![]() Y angle is usually 150-180
Y angle is usually 150-180![]() . Symmetrical O
. Symmetrical O![]() H
H![]() O hydrogen bonds are very strong and are approximately 2.38 Å long (O
O hydrogen bonds are very strong and are approximately 2.38 Å long (O![]() O distance).
O distance).
Van der Waals contacts may be represented by considering van der Waals radii for each atom. These are listed by Pauling in 'The Nature of the Chemical Bond'. H 1.2 Å, C 1.7, N 1.5, O 1.4, P 1.9, S 1.85, F 1.35, Cl 1.8, Br 1.95, I 2.15. Half-thickness of aromatic molecule is 1.85 Å. Radius of methyl group CH3 2.0 Å.
Metal coordination. Metal ions tend to gather negatively charged groups, e.g. the oxygen atoms of carboxylate ions, around them. Distances are shorter than van der Waals distances. The values depend on the number of surrounding oxygen atoms. Li+![]() O 2.0 Na+
O 2.0 Na+![]() O 2.4 K+
O 2.4 K+![]() O 2.8-2.9 NH+4
O 2.8-2.9 NH+4![]() O 3.0 Mg2+
O 3.0 Mg2+![]() O 2.1 Ca2+
O 2.1 Ca2+![]() O 2.4-2.5 Zn2+
O 2.4-2.5 Zn2+![]() O 2.0-2.2 Mn2+
O 2.0-2.2 Mn2+![]() O 2.2 Å.
O 2.2 Å.
d. Stereoviews
Stereoviews are often presented in journals. The reader can either view these with stereoglasses, or else he can focus on the two images until an image between them begins to form and then he can allow his eyes to relax until the central image becomes three-dimensional. This takes practice.
4. Absolute configuration
When a crystal contains a molecule that is not superimposable upon its mirror image (its 'enantiomorph'), the absolute configuration or chirality of this molecule may be determined by X-ray crystallography. This can be done if the crystal contains an atom which absorbs X-rays to an appreciable extent, so that a phase change occurs for the X-rays scattered by that atom (relative to the phase of X-rays scattered by the other atoms in the structure). This is referred to as 'anomalous scattering' and causes intensities of reflections with indices hkl and -h-k-l to differ. It is possible to calculate the intensity difference for a given model, i.e. whether I(hkl) is greater than I(-h, -k, -l) or not. A comparison with experiment tells us if our model has the correct hand or not. If not, then x, y and z in the model under consideration should be reversed in sign.
It is important that the students understand the ways that absolute configuration can be described. These are listed below:
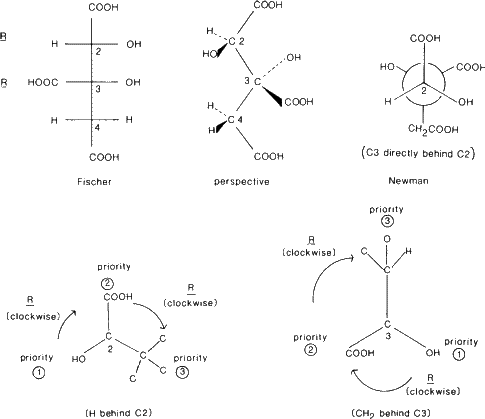
a. R/S system
(Cahn, R. S., Ingold, C. and Prelog, V., Angew. Chem. Int. Ed. 5 (1966), 385).
The 'priority' of an atom is determined by its atomic number. The higher the atomic number the higher the priority. The substituents around a carbon atom are listed in order of their priority. If two or more atoms are identical consider their next substituents, etc. Then look down the bond so that the atom of lowest priority is behind the carbon atom under consideration. If the order of the other substituents going from highest to lowest priority is clockwise the carbon atom is designated R (rectus). If it is anticlockwise it is S (sinister).
b. Fischer formulae
The longest carbon chain is drawn vertically. At each carbon atom under consideration the C--C--C of the main vertical chain is bent so that the top and bottom atoms are below the plane of the paper, and with horizontal X--C--Y angle (where X and Y are substituents) bent up so that X and Y lie above the plane of the paper.
Note: If there is more than one asymmetric carbon atom in the molecule the model must be turned over when the second asymmetric carbon atom is considered. Therefore, the Fischer formula of such a molecule does not look like the molecule in three-dimensional space.
c. Newman projection
A molecule is viewed along the bond between two atoms. The atoms, represented by circles, have bonds drawn to the centre of the upper atom but only to the edge of the circle of the lower atom.
d. Example. (+)-hydroxycitric acid
5. The resolution of the structure
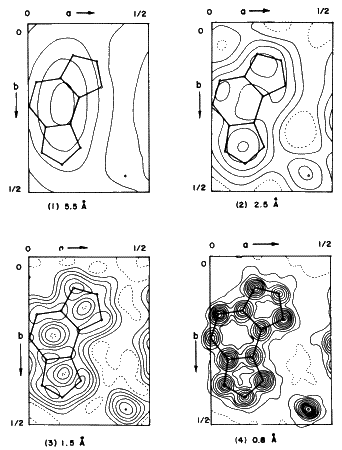
If some of the radiation scattered by an object under examination with a microscope escapes rather than being recombined to form an image, the image so formed will be an imperfect representation of the scattering object. Fine detail will remain unresolved. Similarly, with X-rays, if the diffraction pattern for the customary wavelengths is observed only out to a relatively small scattering angle, the resolution of the corresponding image reconstructed will be low. Furthermore, the resolution will be limited by the wavelength chosen even if the entire pattern is observed. The 'resolution' obtained is usually expressed in terms of the interplanar spacings ![]() ,corresponding to the maximum observed
,corresponding to the maximum observed ![]() values. The effect of changing resolution on the appearance of an electron density map is shown in Fig. 5. Often, with macromolecules, the order does not persist from unit cell to unit cell to high resolution. This lack of high resolution may also be observed when data are collected for crystals near their melting points.
values. The effect of changing resolution on the appearance of an electron density map is shown in Fig. 5. Often, with macromolecules, the order does not persist from unit cell to unit cell to high resolution. This lack of high resolution may also be observed when data are collected for crystals near their melting points.
6. Differences in techniques for small and large molecules
| Smaller molecules | Macro molecules | ||
| 1. | Data collection | Diffract well to high resolution. Generally stable in air. Mount dry on fibre unless unstable. Several thousand reflections. | Diffract to lower resolution (some to higher resolution). Generally unstable to drying. Mount in capillary with mother liquor. Tens of thousands of reflections or more. |
| 2. | Structure solution | Patterson, heavy atom or direct methods. One crystal only. | Isomorphous replacement. This involves many crystals. Heavy atom derivatives required. |
| 3. | Refinement of structure | Full matrix least squares methods. Hydrogen atoms located. | Fitting of polypeptide or polynucleotide chain to electron density map. Constrained and/or restrained least squares methods. Hydrogen atoms not located. Most carbon atoms are also not well-defined except for high-resolution studies. |
See Figs. 3 and 4 (flow charts).
7. Small structures in biology
Many of the important functions of the cell involve small molecules. The list includes substrates or inhibitors of essential enzyme systems, coenzymes, vitamins, antibiotics, steroid hormones and toxic chemicals. A few illustrative examples follow. They will show the techniques used and the types of information that result. Many good examples are not given because of space and time limitations.
a. Uses of cell dimensions and refractive indices
In the early 1930s, before any but the simplest structure analyses were possible, the X-ray crystallographers assisted the chemists by demonstrating that the formula derived for steroids by Wieland and Windaus in 1928 could not be correct since the molecule could not be fit into the unit cell for which dimensions were determined crystallographically. Not only did Bernal, Crowfoot and Fankuchen show that the steroid molecule must be longer and thinner than the formula indicated, but, by considering the maximum and minimum directions of refractive indices, they showed exactly how the steroid molecule must lie in the unit cell, and from this where the functional groups for hydrogen bonding were located in the molecule. Since that time the structures of hundreds of steroids have been determined, verifying these early structural deductions.
References
1. Crowfoot, D., Harris, R. S. and Thimann, K. V. (eds.), Vitamins and Hormones, Vol. II, p. 409, New York, Academic Press, Inc. (1944).
2. Duax, W. L. and D. A. Norton, Atlas of Steroid Structure, Vol. 1, New York, Washington, London, Plenum (1975).
b. Determination of chemical formulae
One particularly useful result of a structure analysis is the determination of the unknown chemical formula of a compound. Three such examples of such determinations solely by crystallographic methods are penicillin which was shown to contain a ![]() -lactam ring system, vitamin B12 which was known to contain cobalt but was shown by X-ray methods to have a modified porphyrin-like structure, and batrachotoxin, a potent poison (used by Colombian Indians for their arrow tips and isolated from a frog) for which only minute quantities could be isolated. Such structure determination then opened the doors for the study of the detailed chemistry (including synthesis), biochemistry and biology of these compounds.
-lactam ring system, vitamin B12 which was known to contain cobalt but was shown by X-ray methods to have a modified porphyrin-like structure, and batrachotoxin, a potent poison (used by Colombian Indians for their arrow tips and isolated from a frog) for which only minute quantities could be isolated. Such structure determination then opened the doors for the study of the detailed chemistry (including synthesis), biochemistry and biology of these compounds.
References
1. Penicillin. Crowfoot, D., Bunn, C. W., Rogers-Low, B. W. and Turner-Jones, A., The Chemistry of Penicillin, Princeton, University Press (1949).
2. Vitamin B12. Brink, C., Hodgkin, D. C., Lindsey, J., Pickworth, J., Robertson, J. H. and White, J. G., Nature 174 (1954) 1169; Hodgkin, D. C., Pickworth, J., Robertson, J. H., Trueblood, K. N., Prosen, R. J. and White, J. G., Nature 176 (1955) 325; Hodgkin, D. C., Kamper, J., Mackay, M., Pickworth, J., Trueblood, K. N. and White, J. G., Nature 178 (1956), 64.
3. Batrachotoxin. Tokuyama, T., Daly, J., Witkop, B., Karle, I. L. and Karle, J., J. Amer. Chem. Soc. 90 (1968) 1917; Karle, I. L. and Karle, J., Acta Cryst. B25 (1969) 428.
c. Studies of flexible molecules to obtain comformational information
Certain flexible molecules with important functions in biology are hard to crystallize. Two such molecules are ATP, the carrier of energy to those cellular processes that require the input of energy, and NAD+, the coenzyme in certain reactions e.g. those of dehydrogenases. These two molecules have flexible pyrophosphate linkages. Such structures, not determined to very high resolution, are hard to study because the crystal quality is generally poor and the data may present difficulties in structure solution. However the general conformation of the molecule and its packing with metal ions are found. Since the molecule is flexible it is presumed that one of several conformers with similar energies has been studied in a particular crystal structure determination. ATP was studied as a sodium salt and NAD+ as a lithium salt. The resolution of each structure determination was about 1 Å. The coenzyme NAD+ was found in an 'extended' form as seen in enzyme complexes, rather than in the 'folded' form.
References
1. ATP. Kennard, O., Isaacs, N. W., Coppola, J. C., Kirby, A. J., Warren, S., Motherwell, W. D. S., Watson, D. G., Wampler, D. L., Chenery, D. H., Larson, A. C., Kerr, K. A. and Riva di Sanseverino, L., Nature 225 (1977) 333.
2. NAD+. Saenger, W., Reddy, B. S., Muhlegger, K. and Weimann, G., Nature 267 (1977) 225.
d. Studies of molecular packing and hydrogen bonding
In addition to the information obtained on conformation when a crystal structure is determined, there is extensive information on intermolecular packing. This information can be used to advantage particularly if the molecule crystallizes with solvent or if several different crystalline forms are studied. For example, a study of biotin and various different crystalline derivatives and analogs was made. Biotin acts as an intermediate carrier of carbon dioxide during the action of certain carboxylating enzymes. It was found that the carbonyl oxygen atom and one ring nitrogen atom formed hydrogen bonds in a fairly constant manner. It was suggested that this observed hydrogen bonding scheme represents that manner by which biotin recognizes bicarbonate.
Reference
Stallings, W. C., Arch. Biochem. Biophys. 183 (1977) 1819.
e. Studies of enzyme mechanism utilizing structures of substrates and inhibitors
An example of the use of structure in determining the stereochemistry of an enzymatic reaction follows. In the case of aconitase, a Krebs cycle enzyme, there are three interconvertible substrates. The structures of these -- citrate, isocitrate and cis -aconitate -- were studied as various salts. From the results it was possible to suggest a mechanism whereby one of these substrates could be converted to each of the other substrates, and the fact that the hydrogen atom removed from citrate was enzymatically incorporated into isocitrate (and vice-versa) was explained. All known features of the stereochemistry of the substrates obtained from crystal structure analyses were preserved. In addition a mechanism whereby fluorocitrate inhibits the enzyme has also been proposed. Studies on the enzyme itself are now in progress.
References
1. Glusker, J. P., J. Mol. Biol. 38 (1968) 149; Accts. Chem. Res. 13 (1980) 345.
2. Carrell, H. L., Glusker, J. P., Villafranca, J. J., Mildvan, A. S., Dummel, R. J. and Kun, E., Science 170 (1970) 1412.
f. Comparisons of structures using the Cambridge Crystallographic Data file
The results of X-ray structure analyses of small molecules are stored on the Cambridge Crystallographic Data file which is updated several times a year. From this data file known structures or portions of structures can be compared in detail. This is much less time-consuming than the older methods of doing an exhaustive literature search and then analysing results via model building or individual calculations. The results can be used to analyse chemical and biochemical reactivity.
References
1. Wilson, S. R. and Huffman, J. C., J. Org. Chem. 45 (1980) 560.
2. Murray-Rust, P., Bürgi, H. B. and Dunitz, J. D., J. Amer. Chem. Soc. 97 (1975) 921.
8. Enzymes and other proteins
Enzymes are polypeptides which use ordinary chemical mechanisms and specific binding interactions to speed up reactions. No mysterious forces need be invoked. Studies of the mechanisms by which enzymes catalyse reactions have been made by both biochemists and crystallographers, and it is when they work together that the most information is obtained. Once the three-dimensional structure has been determined by the crystallographer, further information may be obtained on the mechanism by NMR studies, chemical modification, and by X-ray crystallographic studies of enzyme-inhibitor complexes. The lock-and-key model of Emil Fischer (the enzyme is the lock and the substrate is the key), with some modification, is relevant to the interactions involved.
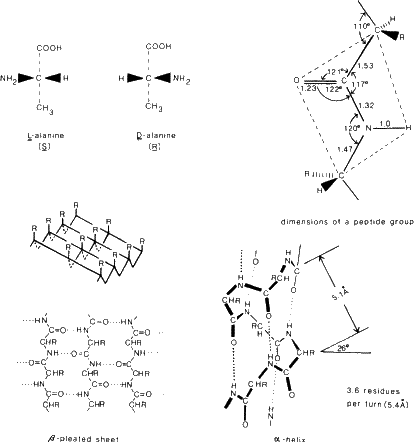
The building blocks are L-amino acids, NH+3--CHR--COO-, where R is a substituent which defines the particular amino acid. There are 20 that are normally found in proteins and the structures of these have been determined by X-ray and neutron diffraction methods. In proteins these amino acids are joined by a peptide linkage C--NH--CO--C. Essentially this is a planar grouping (as found in numerous peptides) and it is hinged to the next peptide group at the carbon atom to give a flexible backbone composed of planar peptide segments. Some dimensions are listed in Fig. 6. However two important types of interactions can occur as the three-dimensional structure of a protein is built up. One is hydrogen bond formation and the other is disulfide bond formation. These interactions stabilize the molecular shape. In particular hydrogen bond formation is responsible for the existence of ![]() -helices and
-helices and ![]() -pleated sheets, so common in enzymes.
-pleated sheets, so common in enzymes.
The main structural features of globular proteins may be generalized as follows (although exceptions are found):
- 1.
- The molecule is very compact.
- 2.
- Most of the polar or hydrophilic R groups lie on or near the surface.
- 3.
- Most of the nonpolar or hydrophobic R groups lie in the interior, hidden from water.
- 4.
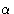 -helices,
-helices, 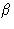 -pleated sheets and disulfide linkages serve to stabilize the structure. In this way various points on the polypeptide chain are brought into close proximity as the polypeptide chain is folded and the active site of the enzyme (i.e. the part of the enzyme where the reaction takes place) is formed.
-pleated sheets and disulfide linkages serve to stabilize the structure. In this way various points on the polypeptide chain are brought into close proximity as the polypeptide chain is folded and the active site of the enzyme (i.e. the part of the enzyme where the reaction takes place) is formed.- 5.
- Proline residues occur principally at bends in the polypeptide chain.
Reference
General stereoviews, Dickerson, R. E. and Geis, I., The Structure and Action of Proteins. Menlo Park, California, Benjamin (1969).
When teaching about macromolecules be sure to look up recent issues of Nature, Proceedings of the National Academy of Sciences, the Journal of Molecular Biology and the Journal of Biological Chemistry for recent advances.
Lysozyme
This enzyme dissolves certain bacteria by cleaving polysaccharides in their cell walls so that the bacterial cells burst. The active site accommodates six sugar residues (ABCDEF) and breaks a bond between sugars D and E. This bond lies near Glu-35 and Asp-52. Glu-35 (non-ionized) donates a proton to C-1 of ring D creating a positive charge there. This positive charge on C-1 (carbonium ion) is stabilized by Asp-52. As a result of protonation the C--O bond is split. Probably ring D is sterically distorted so that it resembles the transition state during the enzyme reaction.
References
Phillips, D. C., Sci. Amer 215 (5) (1965) 78; Blake, C. C. F., Johnson, L. N., Mair, G. A., North, A. C. T., Phillips, D. C. and Sarma, V. R., Proc. Roy. Soc. B167 (1967) 378; Blake, C. C. F., Mair, G. A., North, A. C. T., Phillips, D. C. and Sarma, V. R., Proc. Roy. Soc. B167 (1967) 365; Phillips, D. C., Proc. Natl. Acad. Sci. USA 57 (1967) 493; Imoto, T., Johnson, L. N., North, A. C. T., Phillips, D. C. and Rupley, J. A., in P. B. Boyer (ed.), The Enzymes, Vol. VII, 3rd edition, p. 665, New York, London and San Francisco, Academic Press (1972); Artymiuk, P. J. and Blake, C. C. F., J. Mol. Biol. 152 (1981) 737.
Ribonuclease
This enzyme catalyzes the cleavage of RNA via a cyclic phosphate intermediate. DNA lacks the 2![]() -hydroxyl group essential for formation of this cyclic form. In this enzyme two histidines are located near the bond to be broken. In the cyclization step one acts as a general-base catalyst and the other as a general acid catalyst (His-12 and His-119 in bovine pancreatic ribonuclease). These roles are reversed in the step involving hydrolysis of the cyclic phosphate. The molecule has a well-defined binding cleft. The mechanism seems to involve nucleophilic displacement on phosphorus with a pentacovalent intermediate, with the attacking nucleophile entering opposite the leaving group ('in-line').
-hydroxyl group essential for formation of this cyclic form. In this enzyme two histidines are located near the bond to be broken. In the cyclization step one acts as a general-base catalyst and the other as a general acid catalyst (His-12 and His-119 in bovine pancreatic ribonuclease). These roles are reversed in the step involving hydrolysis of the cyclic phosphate. The molecule has a well-defined binding cleft. The mechanism seems to involve nucleophilic displacement on phosphorus with a pentacovalent intermediate, with the attacking nucleophile entering opposite the leaving group ('in-line').
Reference
Richards, F. M. and Wyckoff, H. W., in P. B. Boyer (ed.), The Enzymes, Vol. IV, 3rd edition, New York, London and San Francisco, Academic Press (1971).
Carboxypeptidase A
This enzyme is a digestive enzyme. It hydrolyses the peptide bond nearest to the terminal carbonyl group in polypeptide chains. The reaction occurs most readily if the carboxyl-terminal residue contains a bulky aliphatic or an aromatic side chain. Esters are also cleaved but by a different mechanism. Zn2+ binds three enzymatic groups (His-156, Glu-72 and His-69) and water. The structure of a complex of the enzyme with glycyl-L-tyrosine (a very poor substrate) has been determined. Substrate carbonyl and a water molecule are aligned between glu 270 and Zn. With peptides the peptide carbonyl group is coordinated to zinc, and displaced water is delivered to the substrate when glu 270 acts as general base. Tyr 248 also forms hydrogen bonds to the substrate and delivers another proton, so that cleavage occurs. When esters are bound, water is not displaced and glu 270 acts by a nucleophilic mechanism to give an anhydride, while Zn--H2O acts as an acid. Tyr 248 is then not required.
References
Lipscomb, W. N., Reeke, G. N., Quiocho, F. A. and Bethge, P. H., Phil. Trans. Roy. Soc. London B257 (1970) 177; Hartsuck, J. A. and Lipscomb, W. N., in P. B. Boyer (ed.), The Enzymes, Vol III, 3rd edition, p. 1, New York, London and San Francisco, Academic Press (1971); Breslow, R. and Wernick, D. L., Proc. Natl. Acad. Sci. USA 74 (1977) 1303; Rees, D. C., Lewis, M., Honzatko, R. B., Lipscomb, W. N. and Hardman, K. D., Proc. Natl. Acad. Sci. USA 78 (1981) 3408.
Hemoglobin
This protein is not an enzyme, but an oxygen carrier. The heme group lies near the protein surface in a hydrophobic pocket. Thus an area of low dielectric constant is provided which favours oxygenation but not oxidation. When oxygen is taken up, the spin state of iron is changed and a small movement of iron occurs relative to the porphyrin ring. This shift is transmitted so that the constraints holding hemoglobin in the deoxy state are relaxed. As a result it is easier for the next subunit to take up oxygen (cooperativity). Neutron diffraction studies have shown that the Fe--O--O angle is 156![]() .
.
Reference
Perutz, M. F., Kendrew, J. C. and Watson, H. C., J. Mol. Biol. 13 (1965) 669; Perutz, M. F., Nature 228 (1970) 726, 734: Padlan, E. A. and Love, W. E., J. Biol. Chem. 249 (1974) 4067; Ten Eyck, L. F. and Arnone, A., J. Mol. Biol. 100 (1976) 3; Baldwin, J. and Chothia, C., J. Mol. Biol. 129 (1979) 175; Shannan, B., Nature 296 (1982) 683.
Chymotrypsin
This enzyme catalyses the hydrolysis of peptide bonds of proteins in the small intestine. It is selective for peptide bonds with aromatic or large hydrophobic side chains (Tyr, Trp, Phe, Met) on the carboxyl side of this bond. Chymotrypsin also catalyses the hydrolysis of ester bonds. X-ray studies have revealed a 'charge relay system' of Asp-102, His-57 and Ser-195. This grouping has been found in a whole group of enzymes called the 'serine proteases'. Neutron diffraction studies on trypsin show that His-57 acts as a base in the catalytic process. The hydrolysis of peptide bonds occurs by general base-catalysed nucleophilic attack on the carbonyl carbon of the substrate by the hydroxyl oxygen of Ser-195. At the same time the hydroxyl proton of serine is transferred to the imidazole of His-57, the chemical base in the hydrolysis reaction.
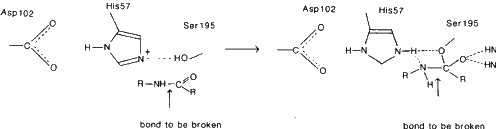
The hydroxyl group of Ser-195 attacks the carbonyl carbon atom of the peptide bond to give a tetrahedral intermediate. His-57 donates a proton to the nitrogen atom of the peptide bond, leading to cleavage and acylation of the enzyme. Deacylation then occurs with water taking the place of the amine group of the substrate.
References
Blow, D. M., Birktoft, J. J. and Hartley, B. S., Nature 221 (1969) 337; Blow, D. M., in P. B. Boyer (ed.), The Enzymes, Vol. III, 3rd edition, p. 185, New York, London and San Francisco. Academic Press (1971); Kossiakoff, A. A. and Spencer, S. A., Nature 288 (1980) 414.
Alcohol dehydrogenase
These are zinc metalloenzymes that oxidize alcohols to aldehydes or ketones. Zinc is bound to the sulphur atoms of Cys-46 and Cys-174 and a nitrogen atom of His-67. An ionizable water molecule occupies the fourth position on the zinc. This water is hydrogen-bonded to the hydroxyl of Ser-48. The oxygen atom of substrate is believed to bind directly to the zinc ion. The nicotinamide ring of NAD+ is bound close to the zinc. In the oxidation of alcohol two hydrogen atoms are removed - one to the 4-position of NAD+ and the other as a proton. The transfer to NAD+ is generally throught to be hydride transfer.
References
Eklund, H., Nordstrom, B., Zeppezauer, E., Soderlund, G., Ohlsson, G., Bowie, T. and Branden, C. -I., FEBS Letters 44 (1974) 200: Eklund, H., Nordstrom, B., Zeppezauer, E., Soderlund, G., Ohlsson, I., Bowie, T., Soderberg, B.-O., Tapia, O., Branden, C.-I. and Akeson, A., J. Mol. Biol. 102 (1976) 27.9. Immunochemistry
An antigen is any object that, when injected into a vertebrate (animal), can stimulate the production of neutralizing antibodies. Immunoglobulins are antibodies. They are Y-shaped proteins, and they bind the antigen and neutralize its effect. This provides a defence against foreign proteins since it allows an organism to distinguish between its own molecules and foreign ones. Potentially immunogenic small molecules are called haptens. The immunoglobulin or antibody has two binding sites so that it can link similar antigens together to form an aggregate that can be destroyed by macrophages.
Each specific antibody possesses a unique sequence of amino acids (in the 'recognition site'). These portions of the antibody fold in three dimensions in a unique way. The antigen causes the synthesis of the specific nucleotide sequence in an RNA template that codes for the desired amino acid sequence. All antibodies are composed of four chains, two light and two heavy and these are linked together by disulfide bonds. Both the light and heavy chain have variable sequences at their amino terminal end and constant regions at their carboxyl ends. The antigen combining sites (the 'active' or 'recognition' or 'binding' sites) are formed by amino acids at the variable regions of both the light and heavy chains.
X-ray studies have given a wealth of information on the three-dimensional structures of the binding sites of two antibodies. The human immunoglobulin protein 'New' that binds vitamin K, and a mouse immunoglobulin protein that binds phosphorylcholine have been studied. These show that residues forming the sites that are complementary to the antigen are contributed by the hypervariable regions (three segments of 5-10 residues each, in both chains). Replacement of amino acids in these areas give rise to binding sites with new specificities, but does not disturb the 'immunoglobulin fold' (the general shape of the immunoglobin).
References
1. Intact human immunoglobulin. Silverton, E. W., Navia, N. A. and Davies, D. R., Proc. Natl. Acad. Sci. USA 74 (1977) 5140.
2. Fab fragment. Poljak, R. J., Amzel, L. M., Zvey, H. P., Chen, B. L., Phizackerly, R. P. and Saul, F., Proc. Natl. Acad. Sci. USA 70 (1973) 3305; Segal, D. M., Padlan, E. A., Cohen, G. H., Rudikoff, S., Potter, M. and Davies, D. R., Proc. Natl. Acad. Sci. USA 71 (1974) 4298: Padlan, E. A., Q. Rev. Biophys. 10 (1977) 35.
3. Fc fragment. Deisenhofer, J., Colman, P. M., Epp, O. and Huber, R., Z. Physiol. 357 (1976) 1421.
4. Immunoglobulin Kol and its antigen-binding fragment. Marquart, M., Deisenhofer, J., Huber, R. and Palm, W., J. Mol. Biol. 141 (1980) 369.
5. Combining region-ligand complex of immunoglobulin NEW at 3.5 Å resolution. Amzel, L. M., Poljak, R. J., Saul, F., Varga, J. M. and Richards, F. F., Proc. Natl. Acad. Sci. USA 71 (1974) 1427.
6. NMR studies using X-ray results. Dwek, R. A., Wain-Hobson, S., Dower, S., Gettins, P., Sutton, B., Perkins, S. J. and Givol, D., Nature 266 (1977) 31.
7. Allergy. Buisseret, Paul D., Scientific American, 247 (1982) 2.
10. Membranes
The bacterium Halobacterium halobium can survive only in solutions with concentrations above 12% salt. When oxygen is in short supply, light is used as a source of energy, first by creating a gradient of hydrogen ions across the cell membrane, and then using this electrochemical gradient to make ATP. Protons are pumped out of the cell. The purple membrane, which occurs as differentiated patches with a regular structure, works as this proton pump and has been studied by diffraction techniques (X-ray and electron diffraction). One molecule of protein, molecular weight 26,000, consists of seven rods 40 Å long (presumably ![]() -helices) extending through the membrane.
-helices) extending through the membrane.
References (purple membrane)
1. X-ray studies of suspension in water. Blaurock, A. E. and Stoeckenius, W., Nature New Biol. 233 (1971) 152.
2. Electron diffraction and structure determination. Unwin, P. N. T. and Henderson, R., J. Mol. Biol. 94 (1975) 425; Henderson, R. and Unwin, P. N. T., Nature 257 (1975) 28.
3. Review. Henderson, R., Ann. Rev. Bioeng. 6 (1977) 87.
Other information on membranes has come from low-angle scattering experiments.
11. Nucleic acids
For an excellent summary of high resolution work on nucleic acid fragments see Dickerson, R. E., Drew, H. R., Conner, B. N., Wing, R. M., Fratini, A. V. and Kopka, M. L., Science 216 (1982) 475. This article contains many useful illustrations. See also Drew, H. R. and Dickerson, R. E., J. Mol. Biol. 152 (1981) 723 and Dickerson, R. E. and Drew, H. R., J. Mol. Biol. 149 (1981) 761.
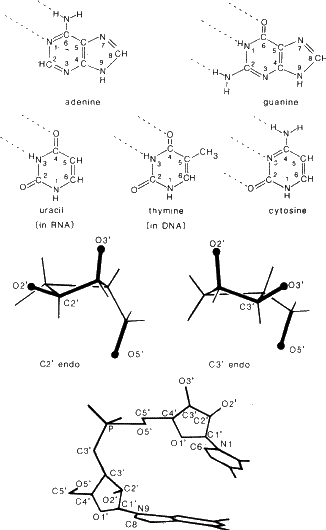
The elucidation of the double-helical nature of DNA was a fundamental step forward in our understanding of the function of DNA. This helicity was deduced from an X-ray diffraction photograph of fibres of DNA taken by Rosalind Franklin (Franklin, R. E., and Gosling, R., Nature 171 (1953) 740; Watson, J. D. and Crick, F. Nature 171 (1953) 737). An analysis of this photograph, expecially if a model of DNA is also available, illustrates some of the principles of diffraction. Since there is the usual reciprocal relationship between distances on the photograph and distances in real space the photograph may be analysed as follows: the very heavy black regions at the top and bottom indicate that the bases stack 3.4 Å apart, perpendicular to the helix axis. The helicity is indicated by the series of diffuse spots in a cross-like pattern in the centre of the photograph. From this follows the distance between points on the helix and its pitch (angle). More recently the double helix has been seen at atomic resolution via the crystal structures of portions of nucleic acids. Thus DNA consists of two polymeric chains twisted around each other in the form of a regular double helix, 20 Å diameter, which makes a complete helical turn every 34 Å. The 'backbone' of the helix consists of sugars linked by phosphate groups. Bases (the pyrimidines, thymine and cytosine, and the purines, adenine and guanine) are linked to the sugars and lie to the centre of the double helix. The two chains are joined by hydrogen bonds between these bases. Adenine is paired with thymine and guanine with cytosine. Thus the two strands are complementary, not identical, and proceed in opposite directions along the helix.
Three major types of helices, A, B and Z have been observed in the structures of small polynucleotides [A (Shakked, Z., Rabinovich, D., Cruse, W. B. T., Egert, E., Kennard, O., Sala, G., Salisbury, S. A. and Viswamitra, M. A., Proc. Roy. Soc. B. 213 (1981) 479), B (Wing, R. M., Drew, H. R., Takano, T., Broka, C., Tanake, S., Itakura, K. and Dickerson, R. E., Nature 287 (1980) 755), Z (Wang, A. H.-J., Quigley, G. C., Kolpak, F. J., Crawford, J. L., van Boom, J. H., van der Marel, G. and Rich, A., Nature 282 (1979) 680)].
The normal form of DNA at high humidity is B-DNA. RNA cannot form a B-helix because the oxygen atom O2![]() interferes with the formation of this. It forms an A-helix. DNA can have both the A and B conformation. RNA-DNA hybrids are always A-type. The B-helix is narrow with no space in the middle, the bases are perpendicular to the helix axis, and there are wide and narrow grooves. In the A-helix the bases are more tilted, there is space in the middle of the helix and the grooves are more nearly the same size. Two conformers of the sugar pucker are found. C2
interferes with the formation of this. It forms an A-helix. DNA can have both the A and B conformation. RNA-DNA hybrids are always A-type. The B-helix is narrow with no space in the middle, the bases are perpendicular to the helix axis, and there are wide and narrow grooves. In the A-helix the bases are more tilted, there is space in the middle of the helix and the grooves are more nearly the same size. Two conformers of the sugar pucker are found. C2![]() endo, found in B-DNA, has C2
endo, found in B-DNA, has C2![]() on the same side of the ring as O5
on the same side of the ring as O5![]() . C2
. C2![]() endo, found in RNA, has C3
endo, found in RNA, has C3![]() on the same side of the ring as O5
on the same side of the ring as O5![]() .Figure 7 shows the formulae of the bases, the types of sugar pucker and the numbering of the polynucleotide. Z-DNA is formed at higher salt concentrations for polynucleotides with alternating purine-pyrimidine sequences, i.e. (AT)n or (GC)n and has the opposite helicity sense to A or B-DNA (i.e. it is a left-handed helix). The Z-DNA helix is long and thin with a very deep minor groove and almost no major groove.
.Figure 7 shows the formulae of the bases, the types of sugar pucker and the numbering of the polynucleotide. Z-DNA is formed at higher salt concentrations for polynucleotides with alternating purine-pyrimidine sequences, i.e. (AT)n or (GC)n and has the opposite helicity sense to A or B-DNA (i.e. it is a left-handed helix). The Z-DNA helix is long and thin with a very deep minor groove and almost no major groove.
Dye molecules, if fairly flat, can intercalate between the bases of DNA and several such structures, using small polynucleotides, have been studied. The phenomenon is well-illustrated by rotating a spring or 'slinky'.
Biology of DNA
1. Replication. When the two strands are separated, the single strands can act as templates for the enzyme-mediated formation of complementary strands.
2. Transcription. The genetic information of DNA is transferred to RNA molecules which act as the primary templates that cause the organization of amino acid sequences in proteins. Complementary base pairs are formed as a DNA strand is copied to give the messenger RNA template for protein formation.
3. Protein synthesis. A specific adaptor, transfer RNA, combines with both the RNA and amino acid, mediating the polymerization of amino acids. Each amino acid has a different transfer RNA which recognizes it and interacts with it by a process called 'activation'. This is effected by a specific enzyme (amino-acyl synthetase), specific for each amino acid. After activation, the amino acid-transfer RNA complex diffuses to the ribosomes where messenger RNA is attached and protein synthesis results.
Transfer RNA
The structure determined for this molecule is L-shaped with anticodon loop, i.e. the part that contains a specific triplet of nucleosides that codes for the particular amino acid for which this is a t-RNA, at one end. At the other end is the amino acid acceptor which recognizes the appropriate amino acid and is esterified in a reaction catalysed by the appropriate aminoacyl synthetase. 60-70% of the structure is double helical. In addition to the above sites for attachment, there is also a site for attachment of the activating enzyme (which binds amino acid to the t-RNA) and for attachment to the ribosome (where transcription occurs). The structure of the amino acyl synthetase is being studied and a suggested model for its interaction with t-RNA has been put forward.
Reference
a. Transfer RNA. Kim, S. H., Suddath, F. L., Quigley, G. J. and Rich, A., Science 185 (1974) 435; Robertus, J. D., Ladner, J. E., Finch, J. T., Rhodes, D., Brown, R. S., Clark, B. F. C. and Klug, A., Nature 250 (1974) 546; Suddath, F. L., Quigley, G. J., McPherson, A., Sneden, D., Kim, J. J., Kim, S. H. and Rich, A., Nature 248 (1974) 20.
b. Tyrosyl t-RNA synthetase. Irwin, M. J., Nyborg, J., Reid, B. R. and Blow, D. M., J. Mol. Biol. 105 (1976) 577.
12. Structures of viruses
Viruses consist mainly of protein and nucleic acid and their organization is of particular interest because they represent a form of life and because they may be crystallized. Two examples, a rod-like virus and a spherical virus, are described here.
Tobacco mosaic virus (TMV) is a rod-like virus 3000 Å long and 90 Å radius with a central hole of radius 20 Å. Identical protein subunits (MW 17,500) form 49 subunits in 3 turns protecting the RNA. Isomorphous replacement was applied to the fibre diagram of TMV. The general conformation could be deduced.
Tobacco bushy stunt virus (TBSV) is a spherical virus that crystallizes in a cubic cell a = 383 Å, space group I23. The virus coat is built from protein subunits having rigid domains connected by a flexible hinger. Each subunit has a binding site for RNA on its inner surface.
Neutron scattering of virus solution is used to obtain low resolution information on viral nucleic acid since H2O-D2O mixtures can be made with scattering powers that match either RNA or protein.
References
1. Virus architecture. Caspar, D. L. D. and Klug, A., Cold Spring Harb. Symp. Quant. Biol. 27 (1962) 1.
2. The assembly of a virus. Butler, P. J. G. and Klug, A., Scientific American 239 (1978) 62.
3. Tobacco mosaic virus. Protein disk. Champness, J. N., Bloomer, A. C., Bricogne, G., Butler, P. J. G. and Klug, A., Nature 259 (1976) 20; Bloomer, A. C., Champness, J. N., Bricogne, G., Staden, R. and Klug, A., Nature 276 (1978) 362.
4. Tobacco bushy stunt virus. 5.5 Å resolution. Winkler, F. K., Schutt, C. E., Harrison, S. C. and Bricogne, G., Nature 265 (1977) 509; Harrison, S. C., Olson, A. J., Schutt, C. E., Winkler, F. K. and Bricogne, G., Nature 276 (1978) 368: Robinson, I. K. and Harrison, S. C., Nature 297 (1982) 563.
5. Neutron small angle scattering. Jacrot, B., Chauvin, C. and Witz, J., Nature 266 (1977) 417.
Acknowledgement
I thank Drs. Michael N. Liebman and Sung Hou Kim for assistance in the preparation of this pamphlet and support from NIH grants CA-10925, CA-06927 and CA-22780.