Proofreading DNA modifications
 Direct chemical editing of DNA might be useful in synthetic biology for modifying genes in an organism for a specific task or in medicine for repairing damaged genes involved in diseases, such as cancer and certain inherited disorders. Unfortunately, as Guillermo Montoya of the Spanish National Cancer Research Centre in Madrid, Spain, and colleagues point out sloppy editing can lead to damage to the genome as a whole.
Direct chemical editing of DNA might be useful in synthetic biology for modifying genes in an organism for a specific task or in medicine for repairing damaged genes involved in diseases, such as cancer and certain inherited disorders. Unfortunately, as Guillermo Montoya of the Spanish National Cancer Research Centre in Madrid, Spain, and colleagues point out sloppy editing can lead to damage to the genome as a whole.
Writing in the journal Acta Crystallographica Section D Biological Crystallography [Stella et al. (2014), Acta Cryst. D70, 2042-2052; doi:10.1107/S1399004714011183] Montoya and colleagues, explain how a new protein-DNA binding domain can allow the necessary modifications to be made due to its high specificity. Their strategy involves allowing specific gene modifications to be made through the addition; removal or exchange of DNA sequences using customized proteins and endogenous DNA-repair machinery.
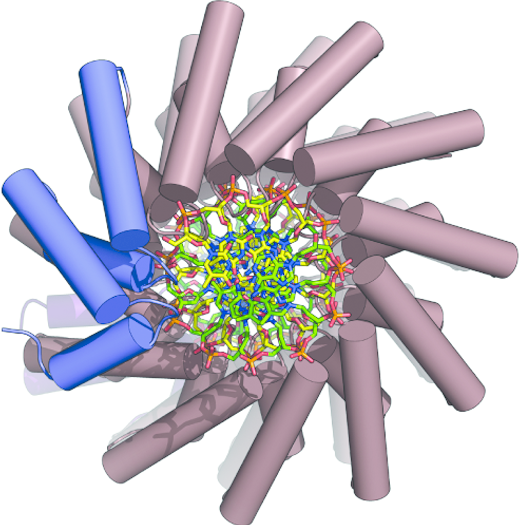
Key to such successful editing will be the engineering of protein-DNA interactions, the team says and this in turn will rely on finding DNA-binding domains. The researchers recently identified one such protein, BurrH, from the bacterium Burkholderia rhizoxinica which contains highly polymorphic DNA-targeting modules. They have demonstrated that BurrH recognizes a 19 base-pair DNA target. Now, they have determined the crystal structure of the apo, unattached, form of the protein and its structure bound to DNA to reveal insights into how form affects function. The team explains that structural data were obtained using a single-wavelength anomalous diffraction (SAD) technique on a selenium derivative (all methionines were substituted for selenomethionines) of the protein crystals chilled to 100 Kelvin to enhance the signals.
The crystal structure uncovered a central region containing 19 repeats of a helix-loop-helix modular domain (BurrH domain; BuD) within the protein's complete chain of 794 amino acids. This molecular recognition unit identifies its DNA target through a single residue-to-nucleotide code. As such, the team reports, this could make it relatively easy to change to allow it to target specific genes. The tailored domains can target specific DNA regions and so by fusing BuD to catalytic domains with DNA-modifying activity the team can shuttle enzymes such as nuclease, methylase etc to specific regions of the genome. In their work Montoya and co-workers show that the process allows targeted mutagenesis to be induced specifically by delivery of the nuclease activity and the insertion of exogenous DNA at the binding site without widespread disruption of the genome.
Indeed, while B. rhioxinica is a symbiont of crop fungi that cause devastating blight in rice, onions and other food crops, the team has now exploited its molecular biology to engineer a protein system based on BuD-derived nucleases (BuDNs) that can target genes in human hemoglobin beta (HBB). The targeting can home in on genes close to the mutations that give rise to sickle-cell anemia and so might ultimately be used to carry out genetic repairs in this debilitating and ultimately lethal disorder.
"Our work shows that this novel platform can be engineered to recognize DNA sequences in human cells," the team explains. They add that it is the combination of efficiency and specificity of BuD as well as the fact that only a single residue, the 13th, need be changed to allow it to target other DNA sequences make it a very useful tool. The researchers add that BuD is "well suited to multiple genome- modification approaches for cell or organism redesign, opening new avenues for precise and safe gene editing for biomedical purposes."


