Synchrotron X-ray footprinting helps visualise water in proteins
 Water plays
a substantial role in the fundamental events of biological processes and
regulates biomolecular structure and dynamics in living cells. Interactions
with water are demonstrated to be critical for protein structure, flexibility
and folding. It has also become increasingly clear that water molecules play an
active role in a number of protein-dependent functions. Because proteins are
the central players in cellular activities it is of vital importance to
understand protein-water interactions.
Water plays
a substantial role in the fundamental events of biological processes and
regulates biomolecular structure and dynamics in living cells. Interactions
with water are demonstrated to be critical for protein structure, flexibility
and folding. It has also become increasingly clear that water molecules play an
active role in a number of protein-dependent functions. Because proteins are
the central players in cellular activities it is of vital importance to
understand protein-water interactions.
X-ray crystallography is one of the tools used to determine the positions of water molecules within proteins at atomic resolution. With the continuous evolution of X-ray sources, detectors, software and protein engineering techniques, very high resolution crystal structures are now routinely available.
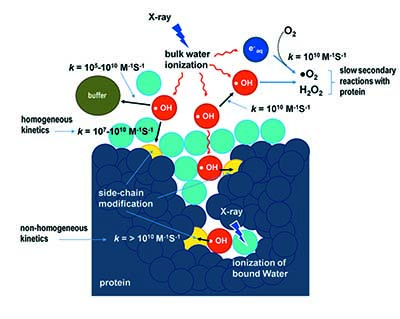
A group of scientists from the US [Gupta et al. (2016), J. Synchrotron Rad. 23. 1056-1069; doi: 10.1107/S1600577516009024] discuss how the method of synchrotron X-ray radiolytic labelling mass spectrometry (XF-MS) can be used to directly distinguish the interactions of bulk and bound water with protein side chains in solution. With this technique, the scientists are able to demonstrate how reactive hydroxyl radicals are generated in situ by high-flux-density X-rays, causing covalent labelling of protein side chains which were detected by mass spectrometry. The in situ covalent labelling approach eliminates most of the complexity associated with sample preparation necessary with other high-resolution techniques and at the same time allows easy comparative structural analysis of protein samples under various physiological relevant conditions.
For the past decade, XF-MS has been integrated effectively with many other structural and biochemical techniques to provide a comprehensive picture of macromolecular complexes and their functional states. The XF-MS method offers a new tool for optimising ligand design by mapping the location and the thermodynamic properties of water molecules at protein binding sites and, at the same time, XF-MS data can be used to develop, validate and refine molecular dynamics approaches that determine water-protein side-chain interactions.
Another exciting future application is the use of X-ray footprinting for protein crystallography, which could yield valuable information in several important areas. Currently dedicated XF-MS beamlines are under development at both the ALS and NSLS II. These beamlines are designed for fully automated sample handling, microsecond irradiation timescales and time-resolved experiments, and will open the door to even further advances in the techniques of XF-MS.


