[Date Prev][Date Next][Thread Prev][Thread Next][Date Index][Thread Index]
--
[Send comment to list secretary]
[Reply to list (subscribers only)]
Ambiguity in atom_site.disorder_group value -1
- To: [email protected]
- Subject: Ambiguity in atom_site.disorder_group value -1
- From: Robert Hanson via coreDMG <[email protected]>
- Date: Mon, 17 Oct 2022 14:38:13 -0500
- Cc: Robert Hanson <[email protected]>
Hello! It has been suggested that I address this issue with this austere group. The issue is that specifying -1 for atom_site.disorder_group has some perhaps unintended consequences.
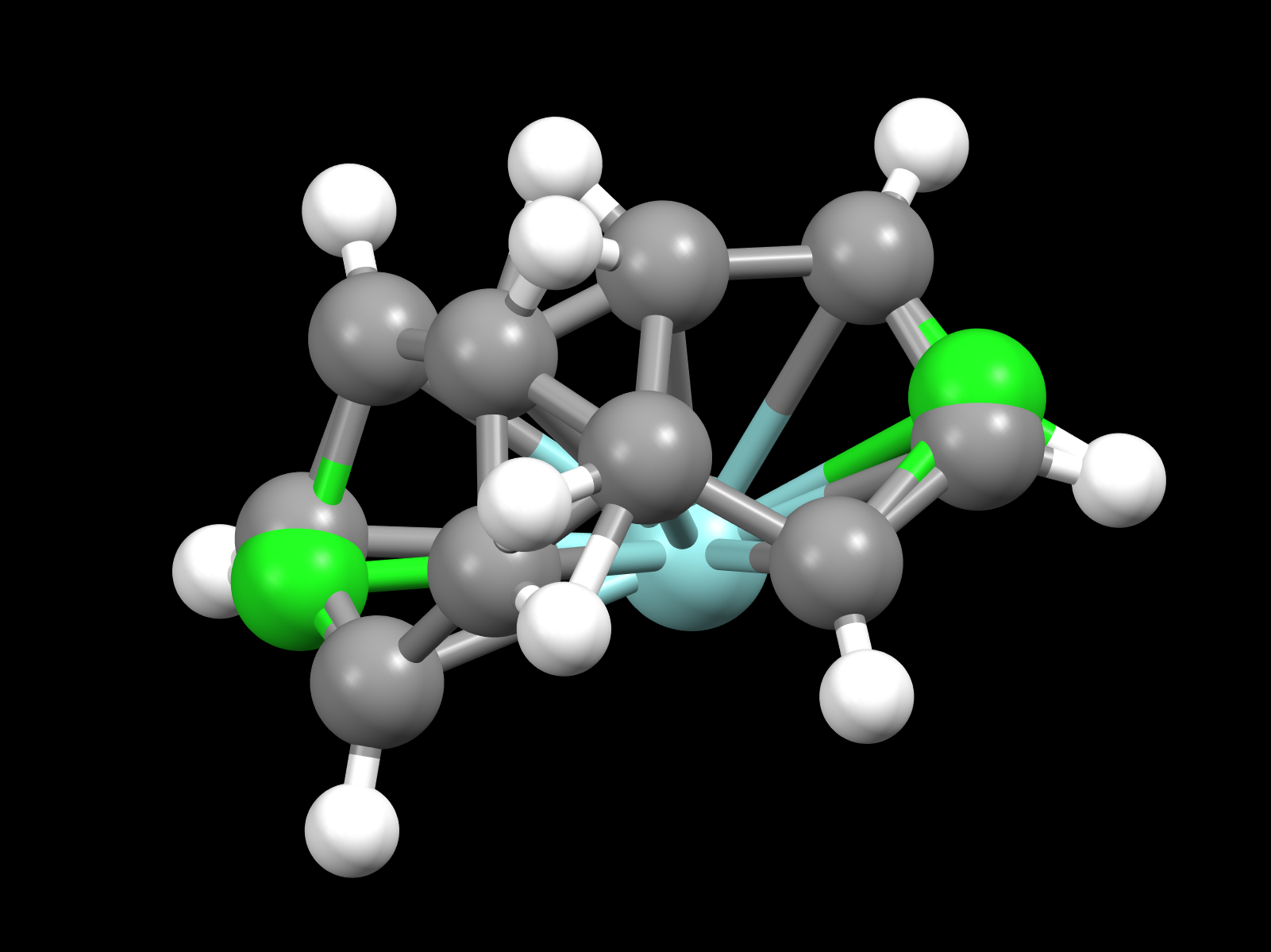
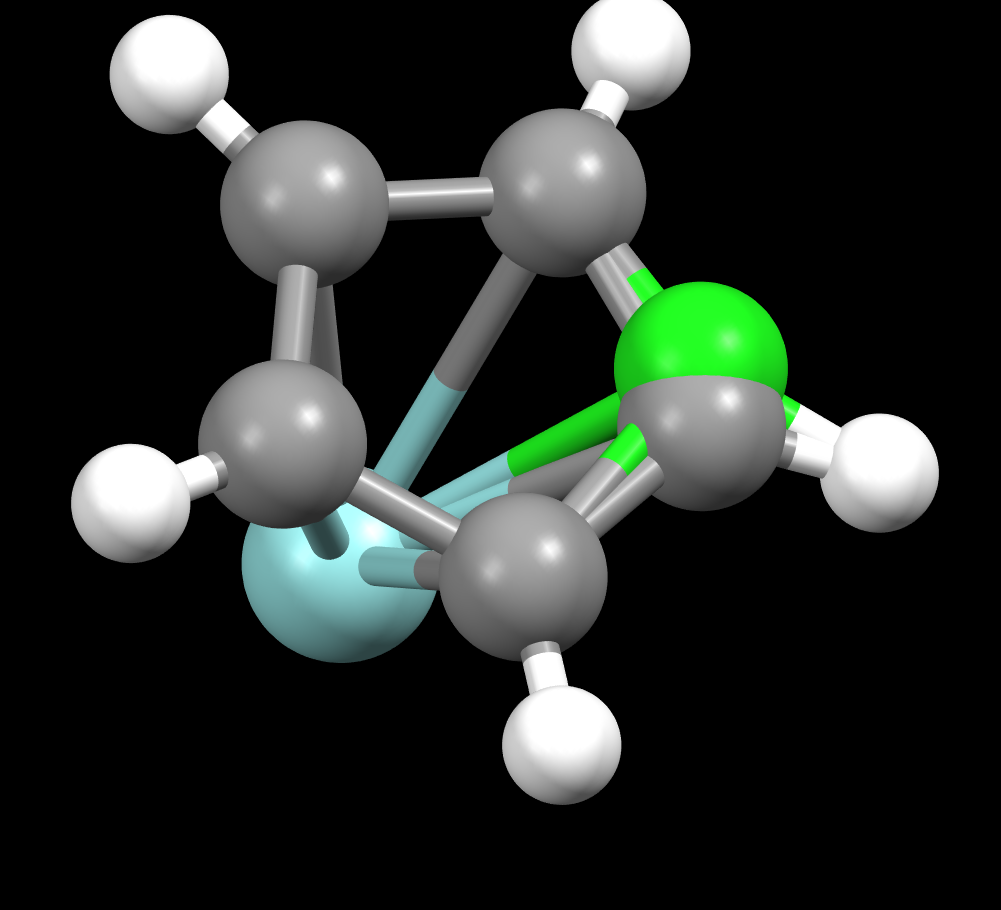
Is it expected (I think it is) that four
copies of the structure are present simultaneously in the actual crystal? It's just that
within each pair we might not know which is present? It's this pairing
aspect that seems to be missing -- if I have the idea right. Certainly the
business about being "about a special position" implies that there is a
special operator, perhaps a C2 axis or an inversion center, that is
considered a key operator. But this is not described in the CIF dictionary description.
The comment we have is this:
A minus prefix (e.g. '-1') is used to indicate sites disordered about a special position.
Brian McMahon helped me understand what that is about. Basically, as I understand it now, sometimes disorder groups are based on space group symmetry. When that happens, rather than using two sets (say) of general positions to describe two different but related disordered groups, only one general position set is required -- applying symmetry then generates the others. Thus, we don't need "group 1" and "group 2"; we only need "group -1".
Visualization software such as Mercury or Jmol should recognize this and refrain from connecting atoms that are in different symmetry-related groups (using whatever distance-related algorithms they have for that). Recently, though, I was working with a CIF file with such a designation (see "disorder-1.cif", attached) and realized that Jmol was handling these incorrectly. I fixed that in Jmol 14.32.77 but also noticed that Mercury 2022.2.0 also had the same issue -- it is connecting atoms across symmetry-related sets:
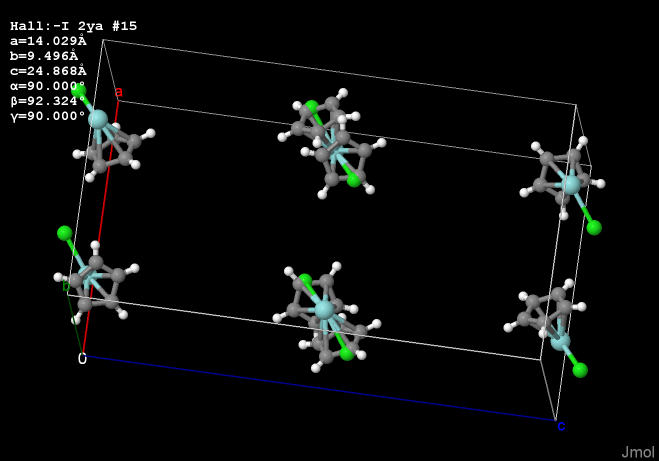
That fixed for Jmol, I realized another issue that is a bit curious. The important question now for me is how the n copies of the symmetry-equivalent
groups are related in a space group with n symmetry operators. For example, in this case, seven "ghosts" are created by applying
all 8 operators in the example I provided.
Brian mentioned that I should offer a change in the description. Here is my suggestion:
A minus prefix (e.g. '-1') is used to indicate that related disordered groups are symmetry equivalent
(for example, involving two orientations related by a C2 axis or an inversion center) and thus
are not listed individually in the atom_site block. In this case, software generating structures
should consider symmetry-equivalent groups derived from these sites to be separately bonded
entities.
As for rendering, I suggest that Mercury has a bug there in terms of bonding. Also with its rendering of the asymmetric unit, which for some reason is using the wrong coordinate for the chlorine atom:
As for Jmol, it is doing this correctly. But in order to do that, it has to create eight independent "alternative locations"
$ print {*}.altloc.pivot
{
"" : 6
"1" : 11
"A" : 11
"B" : 11
"C" : 11
"D" : 11
"E" : 11
"F" : 11
"G" : 11
}
{
"" : 6
"1" : 11
"A" : 11
"B" : 11
"C" : 11
"D" : 11
"E" : 11
"F" : 11
"G" : 11
}
Which, of course, is a bit odd.
Comments?
Bob
--
Robert M. Hanson
Professor of Chemistry
St. Olaf College
Northfield, MN
http://www.stolaf.edu/people/hansonr
If nature does not answer first what we want,
it is better to take what answer we get.
-- Josiah Willard Gibbs, Lecture XXX, Monday, February 5, 1900
Professor of Chemistry
St. Olaf College
Northfield, MN
http://www.stolaf.edu/people/hansonr
If nature does not answer first what we want,
it is better to take what answer we get.
-- Josiah Willard Gibbs, Lecture XXX, Monday, February 5, 1900
We
stand on the homelands of the Wahpekute Band of the Dakota Nation. We
honor with gratitude the people who have stewarded the land throughout
the generations and their ongoing contributions to this region. We
acknowledge the ongoing injustices that we have committed against the
Dakota Nation, and we wish to interrupt this legacy, beginning with acts
of healing and honest storytelling about this place.
_______________________________________________ coreDMG mailing list [email protected] http://mailman.iucr.org/cgi-bin/mailman/listinfo/coredmg
[Send comment to list secretary]
[Reply to list (subscribers only)]
- Follow-Ups:
- Re: Ambiguity in atom_site.disorder_group value -1 (Bollinger, John C via coreDMG)
- Prev by Date: Re: Proposal to define data names for atomic analysis information
- Next by Date: Re: Ambiguity in atom_site.disorder_group value -1
- Prev by thread: New data names to capture the results of atomic composition analysis
- Next by thread: Re: Ambiguity in atom_site.disorder_group value -1
- Index(es):

