Crystallographic databases at International Data Week
Denver, Colorado, USA, 12 September 2016
A session on Crystallography and Structural Data Bases was organised at the SciDataCon meeting during International Data Week to explain to a general audience how some of the world’s foremost research databases maintain and deliver access to the enormous research value of their individual high-quality curated resources.
It also demonstrated, in a time of growing financial pressures, that it is possible to sustain a rich and robust ecology of resources, funded in different ways, with sometimes overlapping content, that delivers maximum value to its target multi-disciplinary communities.
![[speakers]](https://www.iucr.org/__data/assets/image/0015/126204/speakers_p.jpg)
John R. Helliwell, Soorya Kabekkodu, Ian Bruno, John Westbrook, Saulius Gražulis.
The abstracts and links to PDF versions of the individual presentations appear below.
- Overview of the diffraction data deposition activities of the crystallography community
- The Crystallography Open Database - new perspectives
- The Cambridge Structural Database
- 75 Years of the Powder Diffraction File: An Overview of Database Development for Materials Characterization
- Protein Data Bank: An open access resource enabling basic and applied research and education in biology and medicine
Overview of the diffraction data deposition activities of the crystallographic community
John Richard Helliwell, Brian McMahon
Crystallography as a field compares well with other fields such as astronomy and particle physics in ensuring the provision of the underlying data as a formal requirement of research publication, whether it be derived, processed or raw (i.e. primary) data. The raw data archiving is obviously challenging, being the most voluminous. However, this is not the only challenge; the stored raw data’s metadata must be properly described so that it can be assessed and understood. Also, individual raw data sets must be discoverable and reusable by other researchers, whether associated with formal publications or not.
This session at SciDataCon2016 gives an overview of the extant crystallographic databases. This contribution describes the active review and great care which the IUCr and the crystallographic community are taking with respect to their data archiving and data reuse activities. Not least we are conscious of course of the many technological advances in data digital storage and communication now available to all fields in science.
The Crystallography Open Database – new perspectives
Saulius Gražulis, Andrius Merkys, Antanas Vaitkus, Armel Le Bail, Daniel Chateigner, Henry Pilliere, Robert T. Downs, Luca Lutterotti, Peter Moeck, Peter Murray-Rust, Miguel Quirós Olozábal, Werner Kaminsky
Summary
Today's connected world crucially depends on the open availability of data on the Internet. The great success of Wikipedia, PDB (Berman et al., 2012) or open sequence databases like UniProt (The UniProt Consortium, 2015) demonstrates the power of data sharing when it is unhindered by paywalls and copy restrictions.
The Crystallography Open Database (COD) builds on the experience of the open databases and harnesses the power of the community to build an openly-available chemical crystallography database on the net. The COD ingests data in CIF format, validates it according to IUCr dictionaries and quality criteria, and offers consolidated data to COD users, again in a standard CIF format. Currently, the COD contains over 360 thousand records, covering the year span from 1915 to present.
The COD story
Numerous people contributed to the COD (http://www.crystallography.net), and the COD now contains data from several databases such as the AMCSD (Rajan et al., 2006), CrystalEye (Day et al., 2012) or the PCOD (Le Bail, 2005). Most structures published in electronic format are represented in the COD, provided they were available on the Internet freely or were donated by authors or their institutions. Some prominent structures published in paper form were also digitalized and included into the COD.
The COD database stores experimental structures of organic, metal organic and inorganic compounds and minerals. The COD collects the result of all types of crystallographic diffraction experiments (X-rays, electrons, or neutrons diffracted from single crystals and powders). In recent years, however, quantum mechanics computations using DFT and other methods became powerful enough to yield reliable structural descriptions, either ab initio or in conjunction with experimental crystallographic techniques. Such structures are also collected and are stored in a sister database, the TCOD (Theoretical Crystallography Open Database). Since the COD, TCOD and PCOD all use the same CIF framework for data representation, all databases can be searched and processed using the same software tools.
To make the COD simple but efficient several ingredients are crucial. Free/Libre Open Source software (F/LOSS) is at the core of COD development, making it easy to reuse COD data and algorithms. An open data standard – the Crystallographic Interchange Framework – and its ontologies maintained by the IUCr are essential for efficient data exchange (Hall et al., 1991; Bernstein et al., 2016).
The COD maintains stable REST interfaces since its inception (Gražulis et al., 2009; Gražulis et al., 2012). 'You know, cool URIs do not change!' (Berners-Lee, 1998). As a result, the COD is ideally suitable for the 21st century's open, connected world, providing stable links to crystal structures on the Web. The stable setup allows instantaneous reuse of COD data in various sites, and provides a mechanism for data citation using stable, unique COD numbers for every structure.
The COD contains a simple search and retrieve Web interface as demonstrated in Fig. 1. In addition to web search, the COD offers download of all data using various protocols (http, svn, rsync) and querying the database directly using a MySQL client. This gives maximum flexibility in cases where the Web interface is not enough, and permits integration of the COD into other websites or standalone programs.
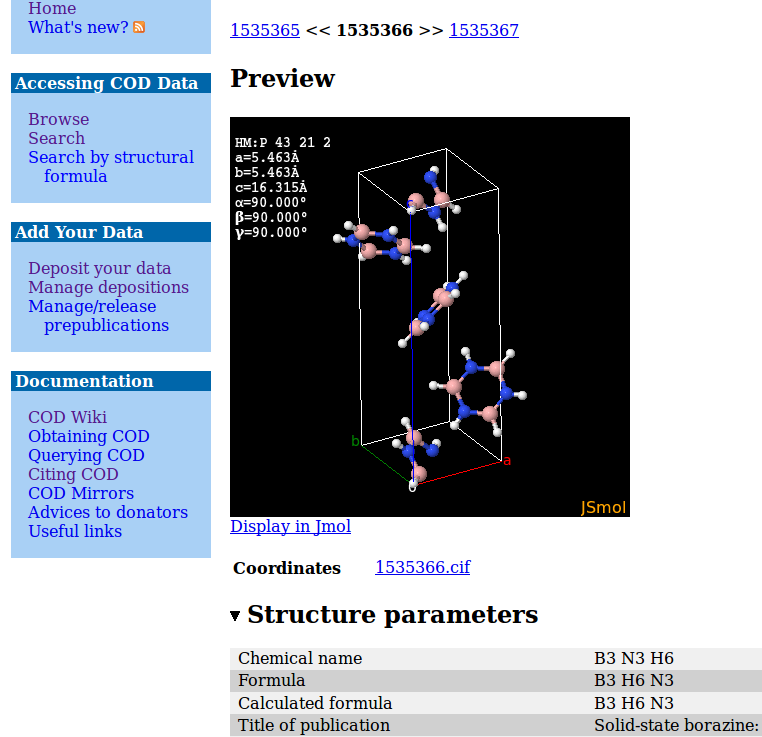
Fig. 1. The COD search result interface and structure view page.
The primary information in the COD is atomic coordinates and crystal descriptions. Moreover, the COD systematically stores Fobs and powder trace data when these are available. COD data entries are manually curated as well as automatically checked. From the ingested data, the COD formulates a new CIF file which is guaranteed to be syntactically correct and contains all original data with attached metadata. Should a change of the COD entry be needed for data curation, the COD meticulously stores all changes in a version control database (currently Subversion), thus providing full traceability and data provenance. With the implementation of these procedures, the COD recently ranked 5 in the Thomson Reuters Databases citation index, and is now recommended by the Nature Publishing Group for all structure depositions.
COD applications
The COD enjoys a growing field of applications. Teaching (Gražulis et al., 2015), powder identification (Lutterotti et al., 2015), source of data for computational material science (First & Floudas, 2013; Pizzi et al., 2016) are among the latest areas where the COD proved useful as a source of data. Moreover, the open nature of the COD allows matching structure descriptions against material properties (Pepponi et al., 2012) yielding the first open collection of material properties and property-structure relations.
In all cases, we benefited from the exisiting crystallographic CIF dictionaries from the IUCr to describe crystallographic entities. We also found it very useful to make domain-specific CIF dictionaries to describe material properties, computational settings and structural metadata. In this way we could reuse the existing CIF dictionaries without duplication, and could reuse our existing software and database structure for new fields of enquiry.
COD new directions
The COD continuity and large collection of crystal data allows us to use it as a data source and management platform for industrial processes: in the recently started SOLSA project, the COD will provide crystal data for mineral identification in mines in real time. Also, the COD will ingest public data that is obtained during such identification runs, further enriching the COD collection and increasing identification precision.
In the year to come, the COD will proceed to store diffraction image data. Storing raw diffraction images currently poses a challenge to meet reasonable management and storage costs, along with enough bandwidth for image set distribution. Here again the COD pursues a community backed, distributed and open-source solution. COD is starting to use a Tahoe-LAFS storage engine (Wilcox-O'Hearn & Warner, 2008), which is currently capable of providing affordable redundant storage up to the order of petabytes.
Acknowledgements
This project has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No 689868 (SOLSA Project).
References
Berman, H., Kleywegt, G., Nakamura, H. &Markley, J. (2012). The Protein Data Bank at 40: Reflecting on the Past to Prepare for the Future. Structure, 20, 391-396. DOI: 10.1016/j.str.2012.01.010
Berners-Lee, T. (1998). Cool URIs don't change. Available at https://www.w3.org/Provider/Style/URI.html [Last accessed 2016-05-29]
Bernstein, H. J., Bollinger, J. C., Brown, I. D., Gražulis, S., Hester, J. R., McMahon, B., Spadaccini, N., Westbrook, J. D. & Westrip, S. P. (2016). Specification of the Crystallographic Information File format, version 2.0. Journal of Applied Crystallography, 49, 277-284. DOI:10.1107/s1600576715021871
Day, N., Downing, J., Adams, S., England, N. W. & Murray-Rust, P. (2012). CrystalEye: automated aggregation, semantification and dissemination of the world’s open crystallographic data. Journal of Applied Crystallography, 45: 316-323. DOI:10.1107/S0021889812006462
First, E. L. & Floudas, C. A. (2013) MOFomics: Computational pore characterization of metal–organic frameworks. Microporous and Mesoporous Materials, 165, 32-39. DOI: 10.1016/j.micromeso.2012.07.049
Gražulis, S., Chateigner, D., Downs, R. T., Yokochi, A. F. T.,; Quirós, M., Lutterotti, L., Manakova, E., Butkus, J., Moeck, P. & Le Bail, A. (2009). Crystallography Open Database - an open-access collection of crystal structures. Journal of Applied Crystallography, 42, 726-729. DOI:10.1107/S0021889809016690
Gražulis, S., Daškevič, A., Merkys, A., Chateigner, D., Lutterotti, L., Quirós, M., Serebryanaya, N. R., Moeck, P., Downs, R. T. & Le Bail, A. (2012). Crystallography Open Database (COD): an open-access collection of crystal structures and platform for world-wide collaboration. Nucleic Acids Research, 40, D420-D427. DOI: 10.1093/nar/gkr900
Gražulis, S., Sarjeant, A. A., Moeck, P., Stone-Sundberg, J., Snyder, T. J., Kaminsky, W., Oliver, A. G., Stern, C. L., Dawe, L. N., Rychkov, D. A., Losev, E. A., Boldyreva, E. V., Tanski, J. M., Bernstein, J., Rabeh, W. M. & Kantardjieff, K. A. (2015). Crystallographic education in the 21st century. Journal of Applied Crystallography, 48, 1964-1975. DOI: 10.1107/S1600576715016830
Hall, S. R., Allen, F. H. & Brown, I. D. (1991). The crystallographic information file (CIF): a new standard archive file for crystallography. Acta Crystallographica Section A, 47, 655-685. DOI: 10.1107/S010876739101067X
Le Bail, A. (2005). Inorganic structure prediction with it GRINSP. Journal of Applied Crystallography, 38, 389-395. DOI: 10.1107/S0021889805002384
Lutterotti, L., Chateigner, D., Pillière, H. & Fontugne, C. (2015). Full-pattern search-match using the Crystallography Open Database: an Internet tool. Available at http://www.ecole.ensicaen.fr/~chateign/danielc/abstracts/Lutterotti_abstract_RXMatiere2013_FPSM.pdf [Last accessed 2016-05-29]
Pepponi, G., Gražulis, S & Chateigner, D. (2012). MPOD: A Material Property Open Database linked to structural information. Nuclear Instruments and Methods in Physics Research Section B: Beam Interactions with Materials and Atoms, 284, 10-14. DOI: 10.1016/j.nimb.2011.08.070
Pizzi, G., Cepellotti, A., Sabatini, R., Marzari, N. & Kozinsky, B. (2016). AiiDA: automated interactive infrastructure and database for computational science. Computational Materials Science, 111, 218-230. DOI: 10.1016/j.commatsci.2015.09.013
Rajan, H., Uchida, H., Bryan, D., Swaminathan, R., Downs, R. & Hall-Wallace, M. (2006). Building the American Mineralogist Crystal Structure Database: A recipe for construction of a small Internet database. In: Sinha A. (Ed.), Geoinformatics: Data to Knowledge, Geological Society of America. DOI: 10.1130/2006.2397(06)
The UniProt Consortium (2015). UniProt: a hub for protein information. Nucleic Acids Research, 43, D204-D212. DOI: 10.1093/nar/gku989
Wilcox-O'Hearn, Z. & Warner, B. (2008). Tahoe: The Least-authority Filesystem. 21-26. Available at https://gnunet.org/sites/default/files/lafs.pdf.
The Cambridge Structural Database
Suzanna Ward, Ian Bruno
Summary
Just over 50 years ago, a small research group based at the University of Cambridge set out to create a compilation of published organic and metal-organic crystal structure data. This group was led by Dr Olga Kennard acting on a vision she shared with the great polymath J D Bernal that “collective use of data would lead to the discovery of new knowledge which transcends the results of individual experiments” (Kennard 1997). In time, this small research group became the Cambridge Crystallographic Data Centre and the initial compilation of data evolved into the Cambridge Structural Database. Much has changed since those early days and this presentation will take a look at how the systems used to maintain the Cambridge Structural Database have had to evolve to accommodate new requirements and how external services have developed to support the continuing quest for new knowledge.
Abstract
The Cambridge Structural Database (CSD) contains the results of over 800,000 crystal structure diffraction experiments and is growing by over 60,000 structures per year (Groom et al. 2016). It is developed and maintained by the Cambridge Crystallographic Data Centre (CCDC). The CCDC provides rich deposition services that promote community norms surrounding publication of crystal structure data and make it easy for a researcher to abide by these. Many structures deposited end up being associated with a journal article but increasingly researchers are using the CSD as their primary platform for publication of their crystal structure data.
A deposited dataset is assigned a Digital Object Identifier (DOI) which resolves to a landing page where the data can be interactively viewed in a web browser and freely downloaded. The CCDC additionally provides mechanisms that enable linking and discovery of data from journal articles and other resources. Rich metadata is associated with each data set using a combination of automated processing and human validation. This includes a chemical representation of the substance studied which is vital for enabling the collection of data to be mined for new knowledge.
The derivation of new knowledge from the data in the CSD is facilitated by a rich set of software applications developed by the CCDC. These enable researchers to search, analyse and visualise the data but also readily apply structural knowledge to a range of disciplines including drug discovery and materials science. As well as enabling research, this area of the CCDC’s activity attracts the financial revenue needed to sustain its core community activities from industry and academia without recourse to direct public funding.
The environment in which the CCDC operates has changed significantly since the early beginnings of the CSD in 1965. The rate at which structures are determined has increased rapidly and the data being deposited has become more complex and more diverse. Changes in the wider economic environment have impacted on our main sources of revenue and thus the services that must be provided to sustain these. General initiatives around research data management and discovery have resulted in opportunities for new services and greater interoperability between resources.
This presentation will focus on how the CCDC has managed its systems over the past half century in order to adapt to the changing environment whilst remaining sustainable. It will look in particular at how internal informatics systems have recently evolved to accommodate increasing throughput and complexity. It will describe how external services are developing to ensure that both data and knowledge in the CSD are widely and appropriately available to a broad range of user communities. It will also reflect on what has been retained from those early days against a backdrop of change. Of particular note here is the validation and review undertaken by expert scientists to ensure that data can be reliably used in the discovery of new knowledge today and in decades to come.
References
Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward S. C. (2016). The Cambridge Structural Database. Acta Crystallographica, B72: 171-179. DOI: http://dx.doi.org/10.1107/S2052520616003954
Kennard, O. (1997). From Private Data to Public Knowledge. In: Butterworth, I. (ed.) The Impact of Electronic Publishing on the Academic Community. London: Portland Press Ltd. pp. 159–166.
75 Years of the Powder Diffraction File™: An Overview of Database Development for Materials Characterization
Soorya N. Kabekkodu, Thomas N. Blanton, Timothy G. Fawcett
Summary
The International Centre for Diffraction Data (ICDD®) Powder Diffraction File is a powerful database for materials identification and has been used extensively by the scientific community for 75 years. Over this period, the database has grown exponentially, housing more than 890,000 diffraction patterns and 330,000 crystal structures in Release 2016. Starting with 1000 hand written file cards in 1941, the Powder Diffraction File (PDF®) has undergone numerous technological developments in the database as well as search/match and data retrieval methods (Faber & Fawcett, 2002). Today, the Powder Diffraction File in Relational Database Format (RDB) format contains extensive chemical, physical, bibliographic and crystallographic data including atomic coordinates enabling qualitative and quantitative phase analysis (Kabekkodu et al., 2002). While using any database in materials characterization, it is important to know the quality of the crystal structure or diffraction pattern found in the database. With varying quality of the published data in the literature, ICDD database editorial review processes had to adopt rigorous data evaluation methods to classify data based on its quality. Every entry in the Powder Diffraction File has a quality mark and editorial comments describing original data errors and any corrections. In addition to X-ray diffraction patterns, the PDF also contains electron and neutron diffraction patterns enabling scientists to use different diffraction techniques to characterize the material of interest. Recent developments in PDF database capabilities and search/match algorithms will be discussed in this presentation.
Development of Powder Diffraction File
The Powder Diffraction File, PDF, started as a collection of X-ray powder diffraction patterns of single phase material providing chemistry, interplanar spacings (d) and relative intensities (I). The amount of data that could be presented was limited in the early days of the Powder Diffraction File as it was published in printed form. In 1969, the Powder Diffraction File was made available to users in the form of magnetic tapes allowing for some additional crystallographic information to be provided. However, data storage was still an issue on account of expensive disk space and slow computer speeds and data retrieval. Today, one can store and access gigabytes of data very rapidly using a personal computer. During continued development in data storage technology, the Powder Diffraction File started adding much more information than just d-I lists to the database. In 2000, the Powder Diffraction File was designed using a Relational Database (RDB) concept which was originally proposed by E. F. Codd (1970). Over the years Relational Database Management Systems (RDMS) have evolved exponentially offering much more robust and efficient database platforms. This facilitated enabling the PDF to store significantly more data including chemical, physical, bibliographic, structural classifications, raw powder patterns, electron and neutron diffraction patterns and crystallographic data including atomic coordinates. All of this additional data content becomes essential when addressing a difficult materials characterization problem. During the past few years ICDD expanded the database scope beyond purely crystalline materials to enable the characterization of amorphous, poorly crystalline and nano materials. The Powder Diffraction File now contains raw diffraction patterns of targeted amorphous, polymer, disordered clays and nano materials. Comparing collected diffraction data with reference raw diffraction patterns is essential for these materials as a d-I list is not sufficient to do a comprehensive phase identification.
In 2011, ICDD extended the design of the database to include modulated structures. As these structures need to be described in 3+d dimensions (super space approach), the PDF database had to be designed in such a way that it does not change the core format that has been used by several search/match software developers for years.
Raw Data Archival
ICDD has been archiving powder diffraction raw data for more than 20 years now. These experimental raw data are stored in the Powder Diffraction File and available for users of the database for their own analysis. Recently the crystal structure of trandolapril was solved by archived powder diffraction raw data in the Powder Diffraction File (Reid et al., 2016)
Quality Marks
Quality marks, assigned to all PDF database entries, are extremely important while working with a large database with multiple experimental determinations. It is important to know the quality of the crystal structure or diffraction pattern found in the database to screen the search/match result set. With varying quality of published data in the literature, database editorial review processes had to adopt rigorous data evaluation methods to classify diffraction data based on its quality. All PDF data are thoroughly reviewed, edited and standardized. ICDD performs more than 100 quality and error checks on the data prior to publication. ICDD’s editorial group has developed an extensive data validation suite to enrich the editorial process. ICDD corrects all of the resolvable errors found in the original data. An entry gets rejected if the magnitude of an error is large and there is insufficient information to correct the observed error. When the quality mark of a PDF database entry is lowered, those entries are populated with editorial comments describing original data errors and any corrections applied. Data validations, corrections, classifications and quality mark assignments are the most important and time consuming step in the editorial process.
This presentation will focus on various aspects of the Powder Diffraction File database emphasizing data validation, classification and search/match methods.
Acknowledgements
The authors would like to thank ICDD members, editors and volunteers for their contributions to the Powder Diffraction File database.
References
Codd, E. F. (1970). A Relational Model of Data for Large Shared Data Banks, Communications of the ACM, 13 (6), 377–387. http://dl.acm.org/citation.cfm?doid=362384.362685
Faber, J. & Fawcett, T. (2002). The Powder Diffraction File: present and future, Acta Crystallogr., Section B: Structural Science, 58, 325-332. http://dx.doi.org/10.1107/S0108768102003312
Kabekkodu, S. N., Faber, J. & Fawcett, T. (2002). New Powder Diffraction File (PDF-4) in relational database format: advantages and data-mining capabilities, Acta Crystallogr., Section B: Structural Science, 58, 333-337 http://dx.doi.org/10.1107/S0108768102002458
Reid, J. R., Kaduk, J. A. & Vickers, M. (2016). The crystal structure of trandolapril, C24H34N2O5: an example of the utility of raw data deposition in the powder diffraction file, Powder Diffraction, available on CJO2016. doi:10.1017/S0885715616000294.
Protein Data Bank: An open access resource enabling basic and applied research and education in biology and medicine
John D. Westbrook
Summary
The Protein Data Bank (PDB) - the single global repository of experimentally determined 3D structures of biological macromolecules and their complexes - was established in 1971, becoming the first open-access digital resource in the biological sciences. It is managed by the Worldwide Protein Data Bank organization. The PDB serves a growing structural biology community, which generates the incoming data, and provides open access for researchers and educators/students around the world to ~120,000 archival entries plus value added information central to basic and applied research and education in the basic and applied biological sciences and medicine. The PDB archive also supports research and teaching efforts in the areas of agriculture, animal husbandry, biological energy production, biochemistry, ecology, protein design, and nanotechnology. The RCSB PDB - the US Regional Data Center for the global Protein Data Bank - currently serves as the PDB archive keeper and is responsible for processing depositions coming from structural biologists based in the Americas and Oceania (deposit.wwpdb.org). The RCSB PDB website (rcsb.org) provides a comprehensive set of "data out" resources with value-added links to other biological data repositories, supporting visualization, comparison, and analyses of the structural data housed in the PDB archive. The importance of open access data sharing and the myriad ways in which interrelationships among science, technology, and the global community have shaped the evolution of the PDB resource for more than forty years will be discussed.
The Worldwide Protein Data Bank
The Protein Data Bank (PDB) is the single global archive for experimentally-determined, atomic-level structures of biological macromolecules. The PDB archive is managed by the Worldwide Protein Data Bank organization, wwPDB; http://wwpdb.org (Berman et al., 2003), which currently includes three founding regional data centers, located in the US (RCSB Protein Data Bank or RCSB PDB; http://rcsb.org), Japan (Protein Data Bank Japan or PDBj; http://pdbj.org), and Europe (Protein Data Bank in Europe or PDBe; http://pdbe.org), plus a global NMR specialist data repository BioMagResBank, composed of deposition sites in the US (BMRB; http://www.bmrb.wisc.edu) and Japan (PDBj-BMRB; http://bmrbdep.pdbj.org). Together, these wwPDB Partners collect, annotate, validate, and disseminate standardized PDB data to the public without any limitations on its use.
The wwPDB Partners are developing the next generation of deposition and annotation tools. A major focus of these tools is validation of submitted atomic coordinates and primary experimental data plus associated metadata. wwPDB Biocurators provide Depositors with detailed reports that include the results of model and experimental data validation. Validation reports were developed and improved using the recommendations from expert Validation Task Forces and Workshops for X-ray (Read et al., 2011), EM (Henderson et al., 2012), NMR (Montelione et al., 2013), and Ligands (Adams et al., 2016). As these wwPDB validation reports provide assessments of structure quality using widely accepted standards and criteria, the wwPDB Partners strongly encourage journal editors and referees to request them from authors as part of the manuscript submission and review process. The reports are date-stamped and display the logo of the wwPDB site where the deposition was curated. They contain the same information, regardless of which wwPDB site processed the incoming data. Provision of wwPDB validation reports is already required by eLife, The Journal of Biological Chemistry, and the International Union of Crystallography (IUCr) journals as part of their manuscript-submission processes. Once each new PDB entry is released, typically at the time of publication, the wwPDB validation reports are made public together with the deposited data.
RCSB Protein Data Bank
To analyze how PDB data relate to other publicly available annotations, the RCSB Protein Data Bank (RCSB PDB, http://rcsb.org, (Berman et al., 2000)) has developed a novel data integration platform that maps 3D structural information across various datasets. This integration bridges from the human genome across protein sequence to 3D structure space.
Users can perform simple searches from the top search bar (e.g. ID, name, sequence, ligand) or build complex combinations of search parameters using Advanced Search. Information from DrugBank is integrated with PDB data to facilitate searches for drugs and drug targets. Other classification systems are used to organize PDB structures in hierarchical trees for browsing and searching (e.g. mpstruc, Gene Ontology, Enzyme Classification).
Protein Feature View provides a graphical mapping of PDB entries onto full-length UniProt sequences. It displays observed and unobserved regions in PDB structures, secondary structure, domain architecture, protein disorder, hydrophobicity profiles, genetic variations, and exon boundaries. Information about available homology models from the Protein Model Portal is also displayed.
Gene View supports visualization of the relationship between genomic and 3D structural data. This tool allows browsing of the human genome with PDB data highlighted onto corresponding genomic ranges. Similar to the Protein Feature View, these data can be correlated with other genomic and protein functional annotations, such as gene structure annotations, DNA repeats, or sequence conservation.
Genome Location to Structure Mapping. A human gene location, e.g. the site of a mutation, can be mapped onto a human gene, and from there to the corresponding protein sequence, and 3D structure. This tool can be used to assess the effect of a Single Nucleotide Variation on the 3D structure.
Validation and Ligand Electron Density Maps. Validation reports and corresponding graphical summary indicators for X-ray, and recently NMR and 3DEM structures are distributed by the wwPDB. These reports assess the quality of each structure and highlight specific concerns. They are provided to Depositors as part of the wwPDB Deposition and Annotation System to help identify potential problems that should be addressed prior to PDB deposition and publication. For each structure, the RCSB PDB displays the graphical validation summary and links to the corresponding report. Also at the RCSB PDB website, 3D visualization of the Sigma-weighted 2m|Fo|-d|Fc| electron density "mini-maps" around ligands is available to assess the quality of ligand modeling.
Acknowledgements
The RCSB PDB is supported by the National Science Foundation (DBI 1338415), National Institutes of Health, and the Department of Energy; PDBe by the Wellcome Trust, BBSRC, MRC, EU, CCP4, and EMBL-EBI; PDBj by JST-NBDC; and BMRB by the National Institute of General Medical Sciences (GM109046).
References
Adams, P. D. et al. (2016). Outcome of the First wwPDB/CCDC/D3R Ligand Validation Workshop. Structure 24(4), 502-508.
Berman, H. M., Henrick, K. & Nakamura, H. (2003). Announcing the worldwide Protein Data Bank. Nature Struct. Biol. 10, 980.
Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). The Protein Data Bank. Nucleic Acids Res. 28(1), 235-42. PMCID: 102472.
Henderson, R., Sali, A., Baker, M. L., Carragher, B., Devkota, B., Downing, K. H., Egelman, E. H., Feng, Z., Frank, J., Grigorieff, N., Jiang, W., Ludtke, S. J., Medalia, O., Penczek, P. A., Rosenthal, P. B., Rossmann, M. G., Schmid, M. F., Schroder, G. F., Steven, A. C., Stokes, D. L., Westbrook, J. D., Wriggers, W., Yang, H., Young, J., Berman, H. M., Chiu, W., Kleywegt, G. J. & Lawson, C. L. (2012). Outcome of the first electron microscopy validation task force meeting. Structure 20(2), 205-14. PMCID: 3328769.
Montelione, G. T., Nilges, M., Bax, A., Guntert, P., Herrmann, T., Richardson, J. S., Schwieters, C. D., Vranken, W. F., Vuister, G. W., Wishart, D. S., Berman, H. M., Kleywegt, G. J. & Markley, J. L. (2013). Recommendations of the wwPDB NMR Validation Task Force. Structure 21(9), 1563-70. PMCID: 3884077.
Read, R. J., Adams, P. D., Arendall III, W. B., Brunger, A. T., Emsley, P., Joosten, R. P., Kleywegt, G. J., Krissinel, E. B., Lutteke, T., Otwinowski, Z., Perrakis, A., Richardson, J. S., Sheffler, W. H., Smith, J. L., Tickle, I. J., Vriend, G. & Zwart, P. H. (2011). A new generation of crystallographic validation tools for the protein data bank. Structure 19(10), 1395-412. PMCID: PMC3195755.
John R. Helliwell and Brian McMahon, Session Organisers

