Workshop on Raw diffraction data reuse: the good, the bad and the challenging
| Organized by | ||
| Loes Kroon-Batenburg (Netherlands), Selina Storm (Germany), John Helliwell (UK) and Brian McMahon (UK) for the IUCr Committee on Data | ||
![[IUCr logo]](https://www.iucr.org/__data/assets/image/0007/61378/IUCr-logo-gradient-RGB.jpg) |
 |
|
Tuesday August 22 2023
Room 205, International Convention Centre, Melbourne, Australia
A major effort of the IUCr Diffraction Data Deposition Working Group (2011 to 2017) and now the IUCr Committee on Data since 2017 has been exploring the practicalities, the costs and benefits, and the opportunities for new crystallographic science arising from large-capacity data archives that have become available. We think it timely to hold a full-day workshop aimed at: (1) discussing current practices in raw data archival and sharing, (2) educating those who generate and deal in crystallographic data on best practices in data reuse in various categories of crystallographic science by leading experts, (3) offering a summing up, including the role of IUCrData’s new Raw Data Letters. We expect attendees to learn about the opportunities for raw data reuse, including the use of raw data as test data sets for machine learning, and to achieve an understanding of how to effectively archive their own raw data to maximise the potential for data sharing and reuse in the future. This workshop will explore in detail the successes and challenges in practice of raw data sharing and reuse. Being a full day, it will complement the proposed microsymposium on "Raw diffraction data reuse: warts and all" in the Congress itself; most importantly the microsymposium will allow for the usual submission of up to four abstracts from anywhere in the world whereas the workshop is principally made up of invited speakers. Furthermore, the microsymposium will highlight the importance of databases. Of the workshop and the microsymposium the proposed keynote on the "European Photon and Neutron Open Science Cloud" (by Andy Götz of ESRF) is the major highlight, making it the world leading effort of this consortium of more than ten European synchrotron and X-ray laser radiation sources with raw data management and sharing.
Remote participation: The session will be accessible remotely via a Zoom meeting link. If you are interested in participating remotely, please register your interest by sending an email to bm@iucr.org. Remote speakers and Chairs are indicated with an asterisk in the programme below.
Reminder: The CommDat user forum is at https://forums.iucr.org
Programme
Times are given in Australian Eastern Standard Time (AEST) (Melbourne) and Central European Summer Time (CEST) (Paris, Berlin).
Tuesday 22 August | Session 1: Facility and raw data archive providers Part IChair: John R. Helliwell |
||
| 08:20-08:30 (00:20-00:30) | Chair | Opening remarks | |
| 08:30-08:50 (00:30-00:50) | Andreas Moll (Australia) | Scientific computing and data management at the Australian Synchrotron | Abstract | Presentation |
|
A. Moll
The Australian Synchrotron is a division within ANSTO and one of Australia’s premier research facilities. It produces powerful beams of light that are used to conduct research in many important areas including health and medical, food, environment, biotechnology, nanotechnology, energy, mining, agriculture, advanced materials and cultural heritage. After 15 years of uninterrupted operation with the original ten experimental end stations, called beamlines, the Australian Synchrotron is currently entering an exciting new phase with the addition of eight new beamlines, including a new high-throughput Crystallography beamline. This created an opportunity for the Scientific Computing team to redesign the whole software stack from the ground up. This presentation will take you on a journey of Scientific Computing at the Australian Synchrotron. You will learn how we employ modern, industry standard tools and architectures in a research environment in order to handle the large data throughput of modern detectors and provide the robustness our users expect from us. A particular focus will be on our use of cloud technologies, running on-premises, across our whole stack from hardware control to data processing on GPUs. (hide | hide all) | |||
| 08:50-09:10 (00:50-01:10) | Anton Barty (Germany) | Managing and curating data flows at PETRA IV | Abstract | Presentation |
|
Anton Barty
The upgrade of the existing PETRA-III synchrotron at DESY to a fourth-generation light source, PETRA-IV, includes not only an increase in brightness but also a new and expanded portfolio of instruments. A good fraction of the planned instruments will generate in excess of a petabyte of data per day during routine operation – a figure that already occurs today at some instruments. At such data rates, retaining all data on disk for 6 months and on tape for 10 years is no longer economically feasible. Instead, rapid analysis using validated pipelines will reduce archived data volumes while providing faster turnaround of results to users performing routine measurements. 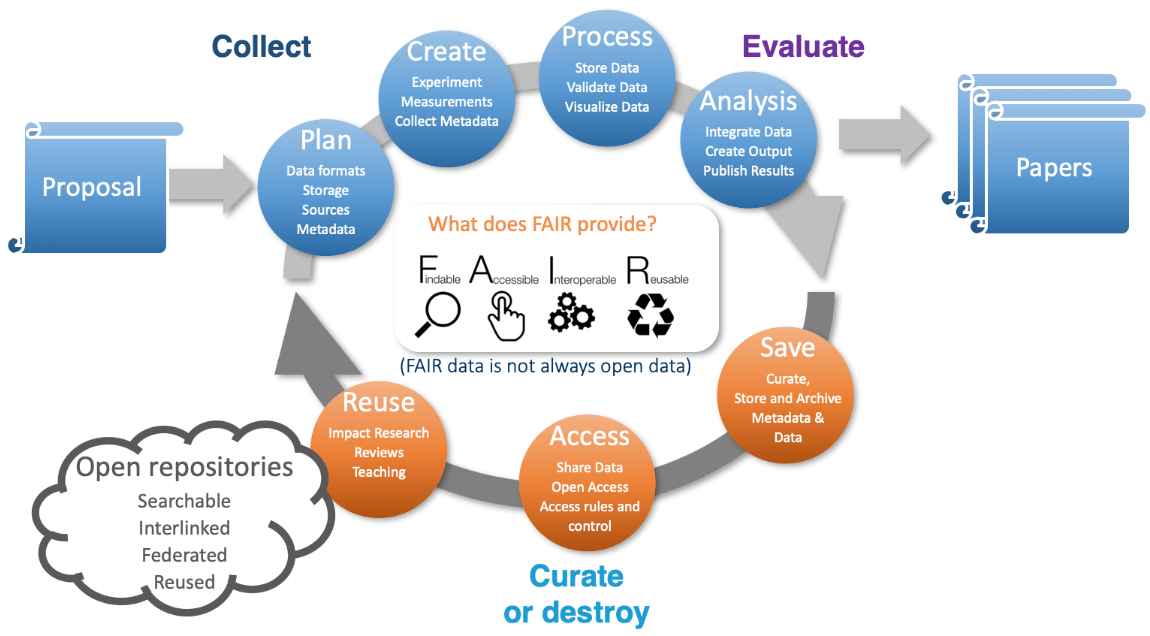 Figure 1. The typical data life cycle at a photon source from proposal through experiment to analysis, curation and open data. The expectation that a majority of users will be experts in their own scientific fields but not necessarily experts in photon science data analysis highlights the need for the provision of high-level integrated data analysis and data management services to users. The PETRA-IV project envisages the provision of analytic services to the wider scientific community, wherein the timely provision of analysed data to users at conclusion of the measurement is essential. With data volumes exceeding the logistical capacity of most users, and especially non-expert users, these services must be provided by the facility or similar large scale research infrastructure. Provision must also be made for commercial measurement services on top of the same core infrastructure where data must be treated in confidence rather than being destined for open publication. Integrated data analysis and data management services are required at the facility to support the full data life cycle from proposal through data taking and on to data analysis, publication, archiving. This includes collection of metadata such as persistent sample identifiers alongside the data, through to eventually making the data open for re-use by the wider community according to FAIR principles (FAIR data stands for Findable, Accessible, Interoperable and Reusable data). Already moving in this direction is the DAPHNE4NFDI national science research data infrastructure project – a cross-institutional project spanning 17 German research institutions currently addressing the topics of data management for photon and neutron science communities. Thanks to Patrick Fuhrmann DESY for the figure, to the DAPHNE4NFDI collaboration, and to the PETRA-IV data team for contributions to the PETRA-IV data management plan. (hide | hide all) | |||
| 09:10-09:30 (01:10-01:30) | * Bridget Murphy (Germany) | DAPHNE4NFDI: DAta from PHoton and Neutron Experiments for NFDI | Abstract | Presentation |
|
Anton Barty[a], Lisa Amelung[a], Christian Gutt[b], Astrid Schneidewind[c], Wiebke Lohstroh[d], Jan-Dierk Grunwaldt[e], Sebastian Busch[f], Tobias Unruh[g], Frank Schreiber[h] and Bridget Murphy[i]
The photon and neutron science community encompasses users from a broad range of scientific disciplines. With the advent of high-speed detectors and increasingly complex instrumentation, the user community faces a common need for high-level, rapid data analysis and the challenge of implementing research data management for increasingly large and complex datasets. The aim of DAPHNE4NFDI [1] is to create a comprehensive infrastructure to process research data from large scale photon and neutron infrastructures according to the FAIR principles (Findable, Accessible, Interoperable, Repeatable). DAPHNE4NFDI brings together users representing key scientific application domains with the large-scale research facilities in photon and neutron science in order to advance the state of data management in the community. The overall goals of DAPHNE4NFDI are:
This work is supported in the context of the work of the NFDI e.V. The consortium DAPHNE4NFDI is funded by the DFG - project number 460248799. [1] Barty, Anton; Gutt, Christian; Lohstroh, Wiebke; Murphy, Bridget; Schneidewind, Astrid; Grunwaldt, Jan-Dierk; Schreiber, Frank; Busch, Sebastian; Unruh, Tobias et al. (2023). DAPHNE4NFDI - Consortium Proposal, Zenodo DOI 10.5281/zenodo.8040605 (hide | hide all) | |||
| 09:30-09:50 (01:30-01:50) | Andy Götz (France) | Making the most of data from the ESRF | Abstract | Presentation |
|
Andy Götz
This talk will present the ESRF approach to data and metadata management for users and the community in general. The latest developments on laboratory information system to provide the right tools to structural biologists to make the most of their data will be presented. Recent developments include developing a flexible but powerful platform for processing data and providing the tools to view and evaluate the results efficiently. The ESRF data repository is being extended to include not only processed data for structural biology but also other domains for example tomography of human organs, fossils and materials science. The talk will conclude with a brief overview of how the ESRF data repository relates to the European PaN data commons and where the next big challenges lie to making data reuse a reality. (hide | hide all) | |||
| 09:50-10:10 (01:50-02:10) | Coffee break | ||
Session 2: Facility and raw data archive providers Part IIChair: * Selina Storm | |||
| 10:10-10:40 (02:10-02:40) | Alun Ashton (Switzerland) | Scientific computing, data sharing and reuse at PSI | Abstract | Presentation |
|
Alun Ashton
The Paul Scherrer Institute (PSI) develops, builds and operates complex large research facilities. The large scientific research facilities at PSI, such as the Swiss Light Source SLS, the free-electron X-ray laser SwissFEL, the SINQ neutron source, the SμS muon source and the Swiss research infrastructure for particle physics CHRISP, offer out-of-the-ordinary insights into the processes taking place in the interior of different substances and materials. These are the only such facilities within Switzerland, and some are the only ones in the world. With new end-stations and detectors at SwissFEL and an upgrade to the SLS due for completion in 2025, the data volumes and computational requirements from photon experiments alone will undoubtedly exceed the current peak of a petabyte of raw data a week. Consequently, with support from initiatives including the ETH Domain program on Open Research Data, the Swiss Data Science Center (SDSC) and the Swiss National Supercomputing Centre (CSCS) and previously from the EU H2020 project ExPaNDS, PSI is already embarking on a holistic approach to handling data and the data lifecycle and finding novel ways to reduce, reuse and share experimental data at each stage of the lifecycle. PSI recently expanded its focus areas and established a new research division: Scientific Computing, Theory and Data (SCD). In recognition of the importance and globally unique ensemble of large facilities at PSI, a keystone to the new division is supporting the operations and experiments with their increasing challenges and opportunities for a unique digital environment. This presentation will focus on the activities of SCD and its collaborators to deliver and improve scientific computing, data sharing and reuse at PSI. (hide | hide all) | |||
| 10:40-11:00 (02:40-03:00) | Genji Kurisu (Japan) | X-tal Raw Data Archive (XRDa): A crystallographic raw diffraction image archive in Asia | Abstract | Presentation |
|
G-J. Bekker[a] and G. Kurisu[a,b]
The Protein Data Bank (PDB) is a public archive of atomic coordinates and crystallographic structure factors including professionally curated meta data. The PDB archive is maintained by the world-wide PDB (wwPDB), a global organization founded in 2003 by RCSB PDB in the USA, PDBe in Europe, and PDBj in Japan, and later jointly managed with the Biological Magnetic Resonance Data Bank (BMRB) and the Electron Microscopy Data Bank (EMDB) as the wwPDB core members [1]. Quite recently, the wwPDB organization welcomed Protein Data Bank China (PDBc) as an Associate member of the wwPDB, and PDBc has started remote processing of some of the structures deposited to and allocated by PDBj. In addition to the PDB core archive, we, PDBj, collaborate with other wwPDB members to maintain the BMRB for experimental data from NMR experiments, and the EMDB for Coulomb potential maps from single-particle or sub-tomogram averaging in Cryo-Electron Microscopy. PDBj is the only wwPDB member who engages in the processing of data for all these three wwPDB core archives [2]. Although the above structural data in the core archives (PDB, BMRB, and EMDB) are actively collected and curated by the wwPDB, the raw image data that were the direct result of the primary experiments, and were used to determine the structures by macromolecular crystallography or cryo-EM microscopy are not collected by the wwPDB. For cryo-EM data, sub-members of the EMDB collect experimental raw micrographs or movies, and archive these in the Electron Microscopy Public Image Archive (EMPIAR). PDBj has been functioning as a local distributor of the EMPIAR archive since 2018, based on a bilateral agreement between EMBL-EBI and Institute for Protein Research, Osaka University. EMPIAR at PDBj (EMPIAR-PDBj) holds the exact same entries as EMPIAR at EMBL-EBI, and we have helped local depositors to transfer their large images/movies, thereby providing our own services (including deposition) through our original website for EMPIAR-PDBj (empiar.pdbj.org/). ![[XRDa home page]](https://www.iucr.org/__data/assets/image/0005/156353/xrda.jpg) Figure 1. The front page of XRDa as operated by PDBj (xrda.pdbj.org). For macromolecular crystallography (MX) raw images, two major archives currently exist; Diamond Light Source in the UK, and the SBDB (SB Grid Data Bank), CXIDB (Coherent X-ray Image Data Bank) and IRRMC (Integrated Resource for Reproducibility in Macromolecular Crystallography) in the USA. However, up till now, no such archive for depositions from Asia has existed, and neither of the existing ones in the UK and the USA are wwPDB members. From 2020, PDBj has started our original diffraction archive named “X-tal Data Archive” (XRDa, xrda.pdbj.org) that securely stores the experimental diffraction images from Asian depositors. As a member of the wwPDB, we have streamlined deposition with the wwPDB’s OneDep system. For depositors from Asia, after depositing their structural data to PDBj via wwPDB’s OneDep system, their entry will be automatically linked to their ORCiD-ID in XRDa. Depositors to XRDa can login using their ORCiD-ID, where any PDB IDs that have been registered in OneDep by them or their co-authors will be available. In addition, depositors can also submit their raw data before submitting their structures to the PDB (and link these afterwards), or submit raw data for structures not to be submitted to the PDB, e.g. for micro electron diffraction data of small molecules. Following login, users can easily deposit diffraction images via the “My entries” page. Once submitted, PDB-linked entries will enter a holding status and will be automatically co-released, while independent entries will be released immediately. Please feel free to deposit your diffraction images to PDBj. XRDa is operated by PDBj and supported by the Platform Project for Supporting Drug Discovery and Life Science Research (BINDS) from AMED under Grant Number JP21am0101066. [1] wwPDB consortium. (2019). Nucleic Acids Research, 47, D520-D528. | |||
| 11:00-11:20 (03:00-03:20) | Fabio Dall'Antonia (Germany) | Handling of big data at the European XFEL | Abstract | Presentation |
|
Fabio Dall’Antonia, Janusz Malka, Egor Sobolev, Philipp Schmidt, Krzysztof Wrona and Luca Gelisio
The European XFEL (EuXFEL) is a unique photon-source facility producing free-electron laser (FEL) pulses in the soft and hard X-ray regime, of extreme brightness and ultra-short duration. These are delivered at MHz repetition rate, enabling various experimental techniques and time-resolved setups. The seven scientific instruments mostly employ pixelized area detectors that can record up to 8,000 1-Mpx images per second. These opportunities for research come at the cost of huge data volumes, which can reach a few PiB per beam-time, posing challenges for data storage and retention, as well as for data re-use purposes. EuXFEL data collected with imagers requires facility services for the correction of pixel intensities. First steps of technique-specific data reduction such as azimuthal integration or crystallographic indexing are done by users remotely on facility resources as well, since download to local computers is not feasible. Currently we are in the process of updating the scientific data policy so as to account for data reduction prior to the long-term storage on disks, as well as for FAIR principles [1] of data management. We are also developing facility services to apply specific data reduction techniques. For example, in case of serial femtosecond crystallography (SFX) [2] data can typically be reduced to only a few percent, sometimes even below 1%, of recorded frames since many FEL shots miss the sample crystals delivered by a liquid jet, leading to images without Bragg diffraction. In this case we are working on facility services that implement Bragg peak detection for automatic filtering procedures, either before data acquisition or at early stages of the offline correction and processing pipeline. Concerning the re-use of open data, EuXFEL has got cloud-based services in the testing stage, which are initially employed for educational purposes but shall become a means for remote data analysis of selected and filtered data sets of each proposal after the embargo period. [1] Wilkinson, M. D., Dumontier, M., Aalbersberg, I. J. et al. (2016). Sci. Data 3, 160018. | |||
| 11:20-11:40 (03:20-03:40) | Wladek Minor (USA) | A subject specific repository for MX (proteindiffraction.org) | Abstract | Presentation |
|
W. Minor, M. Cymborowski and D. R. Cooper
Preservation and public accessibility of primary experimental data are cornerstones necessary for the reproducibility of empirical sciences. We present the Integrated Resource for Reproducibility in Molecular Crystallography (IRRMC). In its first six years, several hundred crystallographers have deposited thousands of datasets representing more than 5,800 indexed diffraction experiments. We will present several examples of the crucial role that original diffraction data played in improving previously determined protein structures. (hide | hide all) | |||
| 11:40-12:00 (03:40-04:00) | Alexandra Tolstikova (Germany) | Processing data in serial crystallography on-the-fly: what kind of raw data do we want to store? | Abstract | Presentation |
|
A. Tolstikova [a], T. A. White [a], T. Schoof [a], S. Yakubov [a], V. Mariani [b], A. Henkel [c], B. Klopprogge [c], A. Prester [c], S. De Graaf [c], M. Galchenkova [c], O. Yefanov [c], J. Meyer [a], G. Pompidor [a], J. Hannappel [a], D. Oberthuer [c], J. Hakanpää [a], M. Gasthuber [a] and A. Barty [a]
Serial crystallography experiments involve collecting large amounts of diffraction patterns from individual crystals, resulting in terabytes or even petabytes of data. However, storing all this data has already become unsustainable, and as facilities move to new detectors and faster acquisition rates, data rates continue to increase. One potential solution is to process data on-the-fly without writing it to disk. Recently, we have implemented a system for real-time data processing during serial crystallography experiments at the P11 beamline at PETRA III. Our pipeline, which uses CrystFEL software [1] and the ASAP::O data framework, can process frames from a 16-megapixel Dectris EIGER2 X detector at its maximum full-frame readout speed of 133 frames per second. The pipeline produces un-merged Bragg reflection intensities that can be directly scaled and merged for structure determination. Processing serial crystallography data on-the-fly offers numerous advantages. It allows for real-time data quality control during the experiment, decreases the time spent between data collection and obtaining a final structure and can significantly reduce the amount of data that needs to be stored and managed. However, there are potential disadvantages and risks to not storing all raw data, such as losing the ability to revisit the data for reanalysis or to reproduce the results. Therefore, careful consideration is needed when deciding which data to store and which to discard. In this talk, we will discuss the challenges and opportunities of real-time data processing in serial crystallography and explore possible strategies for deciding which data to store. [1] White T. A., Mariani V., Brehm W., Yefanov O., Barty A., Beyerlein K. R., Chervinskii F., Galli L., Gati C., Nakane T., Tolstikova A., Yamashita K., Yoon C. H., Diederichs K. and Chapman H. N. (2016). J. Appl. Crystallogr. 49, 680-689. (hide | hide all) | |||
| 12:00-12:40 (04:00-04:40) | Lunch Break | ||
Session 3: Raw data reusers | |||
3.1: Macromolecular crystallographyChair: John R. Helliwell | |||
| 12:40-13:00 (04:40-05:00) | Gerard Bricogne (UK) | The raw, the cooked and the medium-rare: unmerged diffraction data as a rich source of opportunities for data re-use and improvements in both methods and results | Abstract | Presentation |
|
Gerard Bricogne, C. Flensburg, R. H. Fogh, P. A. Keller, I. J. Tickle and C. Vonrhein
Deposition into the PDB of experimental diffraction data, in the form of merged intensities or of `structure factor[ amplitude]s', to accompany atomic models determined and/or refined from them, was made mandatory in 2008. This brought benefits that went well beyond the intended purpose of making the deposited models verifiable and correctable, in the form of an unanticipated `virtuous circle' whereby deposited data fuelled improvements in refinement software that in turn enabled improvements to be made in the initially deposited models, from the same data. The need to manage the outcome of this continuous improvement process led to the introduction of a versioning mechanism into the archiving of atomic models by the PDB in 2017. The creators of the Electron Density Server had already noted in 2004 that `perhaps we should consider deposition of unmerged intensities or even raw diffraction images in the future' [1], without however anticipating the potential for a similar auto-catalytic cycle of simultaneous improvements in data reduction methods and in final structural results that could follow. This potential was later articulated in e.g. [2] in the following terms: `those [merged] deposited X-ray data are only the best summary of sets of diffraction images according to the data-reduction programs and practices available at the time they were processed. Just like refinement software, those programs and practices are subject to continuing developments and improvements, especially in view of the current interest and efforts towards better understanding radiation damage during data collection and in taking it into account in the subsequent processing steps.' Strong general support for the idea of archiving raw diffraction images, together with the recognition that this task was beyond the remit and resources of the PDB, has led over the past decade to the emergence of a delocalised infrastructure (whereby raw data storage and curation takes place at synchrotrons and other dedicated repositories while the PDB provides a capability to annotate entries with a DOI that points to the raw data storage location) that is a major topic in this Workshop. Our own interest has been to document the scientific case for depositing and archiving suitably annotated unmerged diffraction data into the PDB, a goal achievable with modest storage requirements while already creating a standardised resource capable of feeding improvements in scaling and merging methods resulting in better refined models than those originally deposited. This goal is the focus of the current activities of the Subgroup on Data Collection and Processing of the PDBx/mmCIF Working Group of the wwPDB, in which we participate, to expand the mmCIF dictionary to support such extended deposition and archiving. Crucially, unmerged data collected by the rotation method can preserve instrumental metadata about the image number and the detector position at which each diffraction spot was located and integrated, providing a broader decision-making scope over the way it is incorporated into the scaling and merging process. This opens a wide range of possibilities for improving any initially performed scaling/merging steps and for extraction of further data. We will present examples touching upon the following areas:
We are grateful to the PDBx/mmCIF Subgroup on Data Collection and Processing, especially Aaron Brewster, Ezra Peisach, Stephen Burley and David Waterman, for a stimulating collaboration that provided a context for presenting these investigations. [1] Kleywegt, G. J., Harris, M. R., Zou, J. Y., Taylor, T. C., Wählby, A. & Jones, T. A. (2004). Acta Cryst. D60, 2240-2249.[2] Joosten, R. P., Womack, T., Vriend, G. & Bricogne, G. (2009). Acta Cryst. D65, 176-185. (hide | hide all) | |||
| 13:00-13:20 (05:00-05:20) | David Aragao (UK) | Experiences with MX data reuse at Diamond | Abstract | Presentation |
|
D. Aragao, V. Li, S. Collins, G. Winter, R. Gildea, E. Nelson and R. Flaig
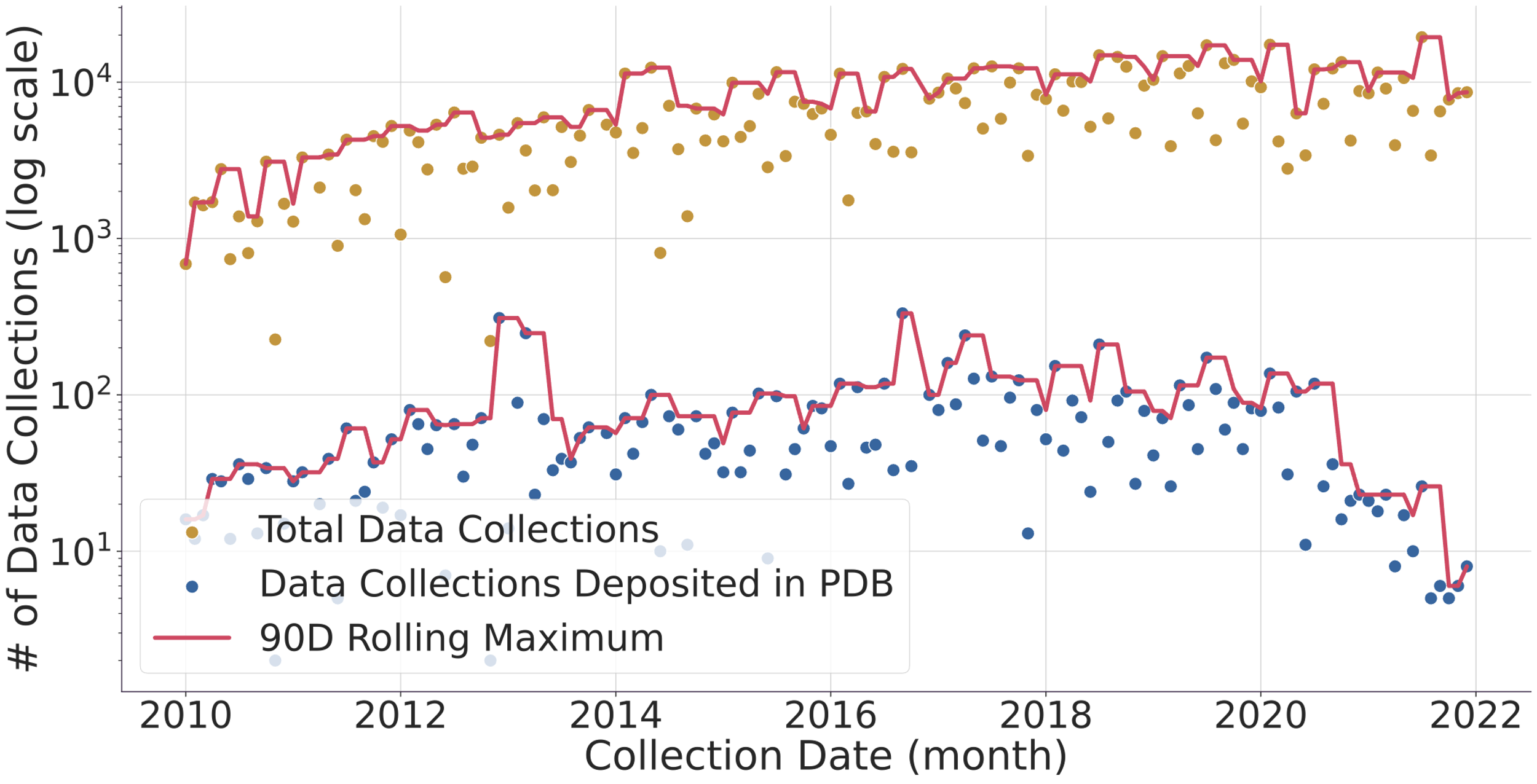 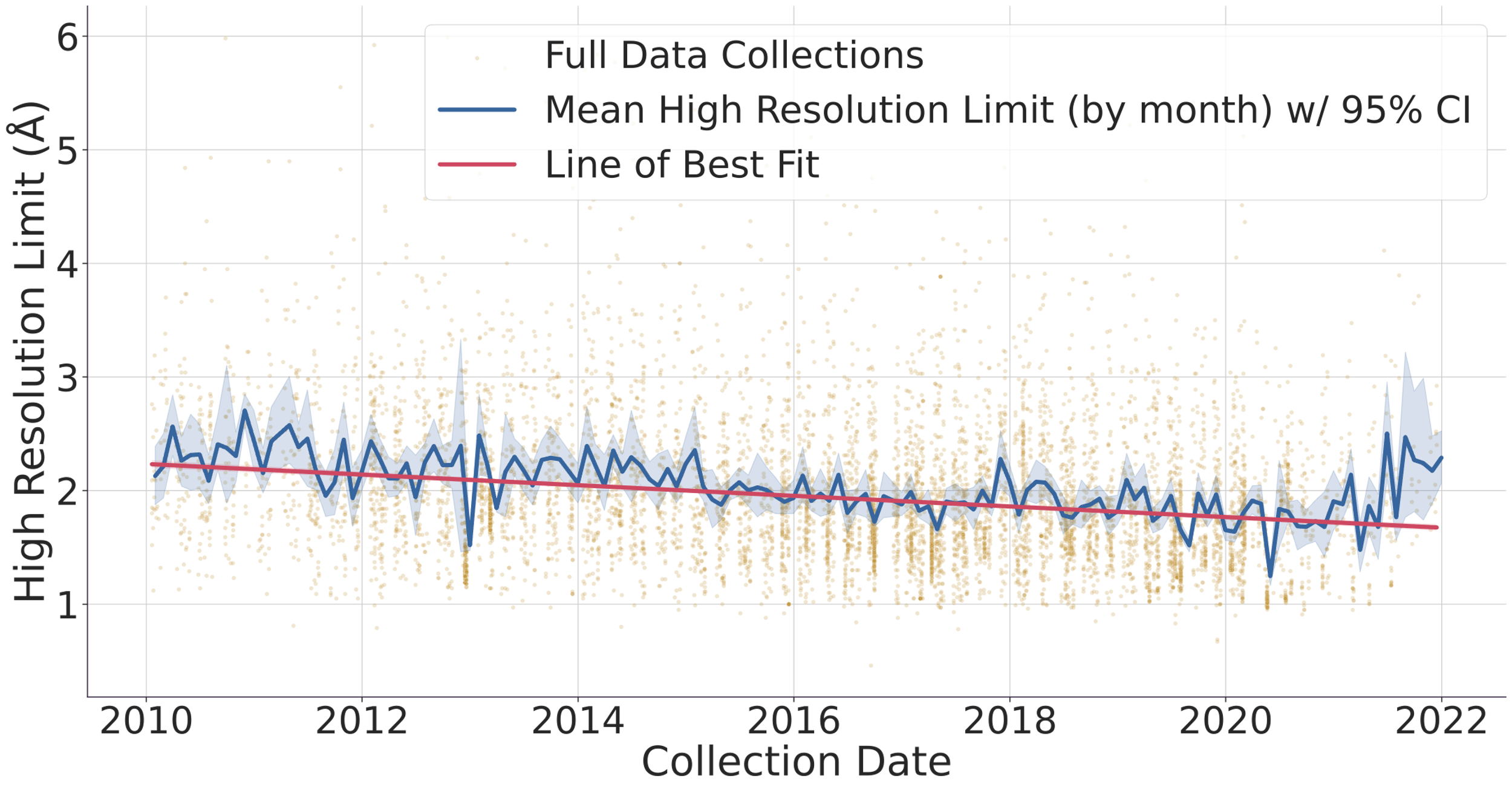 Fig. 1. (top) Evolution of the number of data collected (gold dots) and protein structure depositions (blue dots). Lines represent a 90-day rolling window to smooth the DLS shutdowns where no data is collected. The y-axis of the graph is logarithmic to allow to compare the ~1% deposited structures vs the > 103 datasets collected in the same period. (bottom) Evolution of protein structure high-resolution limit over time with mean and best-fit line. The faded gold dots represent individual full data collections, while the blue line is the average high-resolution limit for each month, surrounded by a 95% confidence interval. The red line is a linear regression line of best fit, and we can see that the high-resolution limit has been getting better over time, improving from around 2.2 ångströms to around 1.7 ångströms. Diamond Light Source (DLS) operates seven beamlines for macromolecular crystallography [1]. These instruments together with others located at similar facilities worldwide record tens of thousands of datasets every week. These are generally left accessible on disk for the industrial partners or academic researchers to analyse. Afterwards, the data are normally stored in a tape system or deleted altogether. With the advent of modern detectors all DLS MX beamlines write HDF5 compressed data and follow the NXmx gold standard [2]. The size of the files and availability of a standard mean these data sets are portable and can be reduced successfully without any extra information. These are critical for data usefulness in the decades to come. On the business end of things, DLS has changed its policy from April 2019 so that academic funded data collected thereafter would be made public from March 2022 making an effective maximum of 3 years embargo with data owners being told 6 months in advance of such. However, DLS has not yet released any data but still has the intention to do it. As for data prior to April 2019, DLS has not yet deleted any data, but such data would be only available at the explicit request of the data owner. To further explore ideas of what can be done with the availability of raw data we present an I04 beamline [3-4] 3-month summer student project where some analysis of the current data in storage, a comparison with the published structures in the PDB and a basic attempt to infer long-term data trends was made. We thank Karl Levy for valuable input and suggestions on how to query the ISPYB database and Diamond Light Source undergraduate summer placement for funding a student at the i04 beamline.
[1] https://www.diamond.ac.uk/Instruments/Mx.html | |||
| 13:20-13:40 (05:20-05:40) | Eugene Krissinel (UK) | Raw data reuse: what it means for CCP4 | Abstract | Presentation |
|
Eugene Krissinel
Collaborative Computational Project Number 4 in Macromolecular Crystallography (CCP4 UK) has a mission to distribute, develop and facilitate development of crystallographic software for all stages of the structure determination pipeline, from raw image processing to phasing, refinement, completion, validation and deposition. Over the 44-year history of the Project, crystallographic software underwent a series of evolutionary changes, caused by advances in theory, sample preparation techniques, quality and properties of raw data. MX is often regarded as a technique with limited reproducibility, which suggests high importance of data retention in the field. For many years, the Protein Data Bank (PDB) collected only end results of interpretation of raw data, the atomic coordinates, leaving no scope for revisiting structure determination in future. The situation improved in 1999 and further in 2020 when, respectively, merged and unmerged data became available for deposition. Deposition of unprocessed, raw data is a natural next step in this direction, which is rather demanding on the storage side and is actively discussed in value for cost terms. We would like to bring the software effect into consideration. Notably, there is no single established format for raw data in MX, and in addition, raw data should be processed with instrument/detector specifics (see Reference [1] as an example). Data processing software, such as XDS [2], HKL [3], Mosflm [4], d*TREK [5], DIALS [6], include extendable sets of routines or plugins for dealing with the variety of formats. Commitment to raw data retention and reusability effectively means commitment to maintaining data processing software, format plugins, and associated beamline metadata forever. This is a significant challenge as software ages faster than data and usually gets retired exactly for maintainability reasons. For example, Mosflm and d*TREK are effectively in sunset mode, and the newest development in the field, DIALS, is not supposed to work with all older formats. Most probably, even if raw data were kept for all PDB entries from day zero, we would not be able to use the oldest datasets today. A possible solution to this problem may be in introducing a “storage” format, but in any case, reuse and storage of raw data cannot be detached from running the corresponding software project. STFC and Diamond synchrotron show an example in maintaining raw data. In 40 days after collection, data are pushed from beamlines to long-term storage, from where they can be downloaded years later. CCP4 is in a good position to facilitate reuse of such data by setting links between data facilities at Diamond, STFC/SCD and CCP4 Cloud [7]. This matches well with introducing CCP4 Cloud Archive facility in January 2023, where completed structure determination projects can be deposited, so that not only the project data and metadata but also the way the structure was solved can be retained; archived projects can be revisited and revised in future. Linking this facility with raw data storage and PDB entry would provide a fully accountable data line for MX. This bears obvious benefits for researchers and makes a foundation for the efficient reuse of collected data, also helping to maximise longevity and robustness of data-handling software. Works have begun in this direction, and much will depend on community take up and feedback. CCP4 is funded by BBSRC UK (Grant BB/S006974/1) and industrial licencing. [1] http://www.globalphasing.com/autoproc/wiki/index.cgi?BeamlineSettings[2] Kabsch, W. (2010). Acta Cryst. D66, 125-132. [3] Minor, W., Cymborowski, M., Otwinowski, Z. and Chruszcz, M. (2006). Acta Cryst. D62, 859-866. [4] Battye, T.G.G., Kontogiannis, L., Johnson, O., Powell, H.R. and Leslie, A.G.W. (2011). Acta Cryst. D67, 271-281. [5] Pflugrath, J.W. (1999). Acta Cryst. D55, 1718-1725. [6] Winter, G., Waterman, D.G., Parkhurst, J.M. et al. (2018). Acta Cryst. D74, 85-97. [7] Krissinel, E., Lebedev, A., Uski, V., Ballard, C. et al. (2022). Acta Cryst. D78, 1079-1089. (hide | hide all) | |||
| 13:40-14:00 (05:40-06:00) | * Melanie Vollmar (UK) | Reusing raw data for machine learning in MX | Abstract | Presentation |
|
M. Vollmar [a] and G. Evans [b,c]
Large quantities of raw diffraction data from protein crystals are collected at synchrotron facilities and in-house X-ray sources every day. The vast majority of this data never yields a protein structure and never leaves the local data storage. Over the last five years there has been a steady increase in interest in the development of machine learning and artificial intelligence models in structural biology. To train any predictive model for high-quality predictions, large quantities of data are required, preferably standardised, curated and labelled. However, the closed state of data storage, i.e. the data is only found on local storage, makes accessing raw diffraction data challenging for anyone who wants to use such data for developing machine learning and artificial intelligence models. Additionally, if raw diffraction data has been made publicly available it usually only represents a certain type of data, namely the one that resulted in successful structure solution, while any diffraction data that did not yield an atomic model remains hidden. Contacting data holders to gain access also often brings the challenge of tracing and finding the raw data on local storage depending on how well data and file management are handled within a facility or research group. Here, we provide a retrospective analysis and share our experiences when developing a machine learning model using raw diffraction data [1]. We describe the challenges in finding suitable data, difficulties accessing that data, efforts needed to trace data locally and, finally, how a well-defined set of raw diffraction data was used to train a machine learning model. We thank Arnaud Baslé, Dominic Jaques, Garib Murshudov, James Parkhurst and David G. Waterman for their contributions and vivid discussions when developing a machine learning model as described in [1]. [1] Vollmar, M., Parkhurst, J., Jaques, D., Baslé, A., Murshudov, G., Waterman, D. and Evans, G. (2020). IUCrJ, 7, 342-354. (hide | hide all) | |||
| 14:00-14:20 (06:00-06:20) | Tea break | ||
3.2: Chemical crystallographyChair: Loes Kroon-Batenburg | |||
| 14:20-14:40 (06:20-06:40) | Jim Britten (Canada) | Use of raw data for diffraction space visualization: What are we missing in an integrated HKL file? | Abstract | Presentation |
|
Jim Britten
For many years chemists, biochemists and physicists have been using area detectors to collect 3D diffraction data from crystals. Engineers are collecting 3D data for texture and residual stress analyses. Various software packages analyse the frames and the derived data is used to solve the problem at hand. ![[Diffraction pattern from nanowire]](https://www.iucr.org/__data/assets/image/0009/156474/nanowire.jpg) Figure 1. Diffraction from a highly oriented nanowire film on a single crystal substrate. In this presentation we will use 3D reciprocal space visualization software [1] to look carefully at examples of these types of data. Very often we can find evidence of unexpected features that have been ignored or missed by the data processing software. In some cases, we can do further processing of the data based on the new-found information. In other cases, we need to flag the data set as a candidate for future analysis when the appropriate software becomes available. It may even trigger the development of new software. Diffraction space is more complicated than we often imagine and contains unmined information about our samples. Scattering from aperiodic crystals is an obvious example. The value of preserving raw 3D diffraction data cannot be underestimated. [1] MAX3D reciprocal space visualization software package (https://rhpcs.mcmaster.ca/~guanw/max3d/MAX3D-5.0-2021.10.12.zip) (hide | hide all) | |||
| 14:40-15:00 (06:40-07:00) | Simon Coles (UK) | The increasing diversity of small molecule data: can one size fit all? | Abstract | Presentation |
|
Simon J. Coles
Today's accepted approaches to handling chemical crystallography data have largely been established in the ‘boom period’ of crystal structure analysis, that is the late 1990s and early 2000s as CCD area detectors took hold and data volumes increased significantly. Chemical crystallography is facing another change, with a range of alternative structure determination methods becoming viable and dynamic crystallography seeing more widespread use. Some examples of initiatives from our laboratory illustrate the nature of this imminent expansion in our field. 3D-Electron Diffraction (3D-ED) is set to significantly impact on small-molecule crystal structure analysis with the introduction of new dedicated instrumentation that will dramatically increase the volume of results generated and lift the technique from research in itself, to being generally applicable and providing a widespread service. 3D-ED is generating some truly amazing results, producing structures from nano crystallites traditionally considered as powders, on materials that normally would never have been applicable to single-crystal analysis. However, the nature of the experiment is challenging and invariably datasets from several crystallites must be merged to maximise the completeness of data, with the result that structures do not meet the same quality standards as we have come to expect from X-ray single-crystal analysis. Other similarly emergent structure determination techniques applied to particular problems, such as NMR Crystallography and XFEL studies, present wonderful opportunities but come with the same data quality problem. The Crystal Sponge technique enables the uncrystallisable to be crystallised. Compounds that do not crystallise well, or at all, or that can only be synthesised in minute amounts can be soaked into a crystalline porous material and the composite host+guest structure determined. This provides a molecular structure that can have great value for synthetic chemists for characterisation or confirmation of product. However, the experimental technique is variable in that molecules can arrange in the sponge in different ways significantly affected by soaking conditions and this leads to diffuse diffraction, disorder and lower quality results. Similarly dynamic crystallography, that is structures under change mediated by e.g. temperature, pressure, gas adsorption, electric current, also suffers from these effects. These exciting advances are set against the backdrop of traditional X-ray crystal structure analysis, with >100 years of enhancing instrumentation, >50 years of collecting results into databases, 40+ years of trusted common refinement processes, 30 years of standards and 20+ years of validation tools. So, the established processes, metrics, etc. for small-molecule crystallography provide a well-established and trusted ‘quality framework’ for our results. This means that the small-molecule crystallography community now caters very well for the validation and quality control of relatively routine structures as part of the checking and publication process. However, this quality framework doesn’t cater well for these exciting new frontiers of chemical crystallography in the sense that results are deemed to be of a lower quality. But the results of these experiments drive and underpin investigations in ways that could never have happened before and with a comparable accuracy to the gold standard of single-crystal X-ray analysis. Being able to answer questions such as ‘what is the compound I have made?’, ‘what is this reaction by-product?’, ‘how has my structure changed?’, ‘how does this material manifest these properties?’, particularly for materials that are not ideally crystalline, can be crucial to further the progress of research. Clearly these are strong examples that extend the community discussions around making raw chemical crystallography data available [1]. But how can we balance this current contrast between well established and emergent techniques? Firstly, it is necessary to consider extending the current quality framework and secondly it is imperative to make raw data available alongside the results from these new techniques. This talk will present the concept of ‘structure grading’ as an indicator of what claims can be made based on a particular result. These claims, and therefore the structure grading, should be backed up by the raw data – particularly in the case of emergent techniques where it is highly likely that methods will improve and so a better result can be derived in the future from the original data. The talk will therefore also consider how it can be shown that the best possible result has been obtained from the raw data, or indications can be provided that declare that there is room for future improvement. [1] When should small molecule crystallographers publish raw diffraction data (2021). Twenty-Fifth Congress and General Assembly of the International Union of Crystallography, https://www.iucr.org/resources/data/commdat/prague-workshop-cx. (hide | hide all) | |||
3.3: Powder diffractionChair: Loes Kroon-Batenburg | |||
| 15:00-15:20 (07:00-07:20) | Nicola Casati (Switzerland) and * Elena Boldyreva (Russia) | Powder diffraction raw data | Abstract | Presentation |
|
Nicola Casati[a] and Elena V. Boldyreva[b,c]
Powder Diffraction is a valuable tool when studying a statistical number of objects, e.g. in quantification or probing chemical reactions or phase transitions. It also provides information on many important compounds and materials that cannot be prepared as single crystals, or that, while starting as single crystals, become polycrystalline due to structural transformations on variations of temperature, pressure, irradiation, etc. Data analysis for powder diffraction is normally not only less straightforward then for single crystals, but is also more prone to ending in false minima and getting wrongly interpreted. A more delicate and possibly robust approach is needed to retrieve the correct information. It is therefore desirable, at times, to go back to original data and find out the origin of specific features in the data themselves, whether coming from the setup, from mishandling or from the experiment itself. This helps not only avoiding mistakes but also extracting the maximum information, which at times can only be viewed a posteriori after more knowledge on the system has been acquired. In this context correct data handling is pivotal to being able to successfully look back at 'old' but still valuable patterns. In the present contribution, we aim to define the important aspects in regulating powder diffraction raw data, including the level at which 'raw is raw' and the metadata that needs to be included, to make sure enough information is kept. This, to ensure it is possible to look back successfully at previous data and/or correct problematic behaviour. The discussion will involve both 1D and 2D detector datasets, including very large ones (several thousand patterns). As one of the examples, matching several aspects, we use a high-pressure study of phase transitions in L-Serine. We illustrate how the raw data collected in 2006 [1] could be used almost 10 years later, in order to get new information from these data, viewed through the new experience obtained in 2015 [2]. EVB acknowledges funding from the Ministry of Science and Education RF (project AAAA-A21-121011390011-4). [1] Boldyreva, E. V., Sowa, H., Seryotkin, Y. V., Drebushchak, T. N., Ahsbahs, H., Chernyshev, V. & Dmitriev, V. (2006). Pressure-induced phase transitions in crystalline L-serine studied by single-crystal and high-resolution powder X-ray diffraction. Chemical Physics Letters, 429(4-6), 474-478. | |||
| 15:20-15:40 (07:20-07:40) | Miguel A. G. Aranda (Spain) | Powder diffraction data sharing and reuse: advantages and possible practical obstacles | Abstract | Presentation |
|
Miguel A. G. Aranda
Scientific data in our crystallographic community can be classified, in broad terms, in three large categories: raw, reduced and derived data. On the one hand and for decades, IUCr has been and is being very active in promoting the sharing of reduced and derived data in independently verified databases. The final results, in narrative style, are also shared in the scientific journals. On the other hand, the need for raw data sharing is clearly increasing, being nowadays technically feasible and likely cost-effective. Within the crystallography field, the powder diffraction (PD) community is a subgroup dealing with several goals, mainly (1) average crystal structure determination; (2) quantitative phase analyses; (3) microstructural analyses; and (4) local structure determination and quantitative analyses of nanocrystalline materials. It should be noted that many PD users are not directly associated with crystallography but with material science, solid-state chemistry and physics, etc., some practices being different in different fields. For PD, derived data for objectives (2) and (3) and to a large extent (4) cannot be incorporated in current `standard' (independently verified) databases. Therefore, and in my opinion, the need for sharing raw PD data is even more compelling than that of sharing raw single-crystal diffraction data. In order to ensure that raw powder diffraction data sharing is useful, the methodology has to be robust. From the computing point of view, the shared data must be findable, accessible, interoperable and reusable – i.e. comply with FAIR standards. However, this is necessary but not sufficient. On the other hand, and from the involved scientific community point of view, the shared data must have sufficient quality and their quantitative reuse should be relatively easy. Some possible benefits of sharing powder diffraction raw data were discussed in a previous publication [J. Appl. Cryst. (2018), 51, 1739-1744]. In this communication, I will further elaborate on the benefits but mainly on some practical obstacles to be addressed. For powder diffraction data from point detectors, the sharing seems to be straightforward. However, for powder diffraction data taken from 2D detectors, this is not the case. It is noted that both correction and integration steps have choices that need to be unified. This is a challenging task that needs to be undertaken. (hide | hide all) | |||
| 15:40-15:55 (07:40-07:55) | Loes Kroon-Batenburg (Netherlands) / * Selina Storm (Germany) | Summing up: the role of IUCrData’s new Raw Data Letters in serving all the above | Raw Data Letters (LKB) |
| 15:55 (07:55) | Close | ||
The Congress Opening Ceremony is at 6 pm | |||


![[A. Moll]](https://www.iucr.org/__data/assets/image/0005/156425/moll.jpg)
![[A. Barty]](https://www.iucr.org/__data/assets/image/0008/156473/barty.jpg)
![[B. Murphy]](https://www.iucr.org/__data/assets/image/0006/156876/murphy.jpg)
![[A. Goetz]](https://www.iucr.org/__data/assets/image/0007/156868/Andy-Gotz.jpg)
![[A. Ashton]](https://www.iucr.org/__data/assets/image/0009/156753/ashton.jpg)
![[G. Kurisu]](https://www.iucr.org/__data/assets/image/0006/156354/kurisu.jpg)
![[F. Dall'Antonia]](https://www.iucr.org/__data/assets/image/0011/156476/dall-antonia.jpg)
![[W. Minor]](https://www.iucr.org/__data/assets/image/0005/113756/minor.jpg)
![[A. Tolstikova]](https://www.iucr.org/__data/assets/image/0003/156477/tolstikova.jpg)
![[G. Bricogne]](https://www.iucr.org/__data/assets/image/0006/156066/bricogne.png)
![[D. Aragao]](https://www.iucr.org/__data/assets/image/0004/156802/aragao.gif)
![[E. Krissinel]](https://www.iucr.org/__data/assets/image/0004/156478/krissinel.jpg)
![[M. Vollmar]](https://www.iucr.org/__data/assets/image/0005/156479/vollmar.jpg)
![[J. Britten]](https://www.iucr.org/__data/assets/image/0010/156475/britten.jpg)
![[S. Coles]](https://www.iucr.org/__data/assets/image/0003/143778/coles.jpg)
![[N. Casati]](https://www.iucr.org/__data/assets/image/0011/156854/casati.jpg)
![[E. Boldyreva]](https://www.iucr.org/__data/assets/image/0010/156853/boldyreva.png)
![[M. Aranda]](https://www.iucr.org/__data/assets/image/0020/143048/aranda.jpg)