


Feature article
30 years of CIF
![CIF_snowball.jpg [Snowball in South Park, Oxford: Kamyar Adl https://www.flickr.com/photos/kamshots/384814496]](https://www.iucr.org/__data/assets/image/0011/152759/CIF_snowball.jpg)
Photograph by Kamyar Adl licensed under Creative Commons CC-BY-2.0
A new paradigm in data characterization
Thirty years ago, in November 1991, Acta Crystallographica Section C published the first of a new category of articles, Regular Structural Papers. At the same time, an article appeared in Acta Crystallographica Section A describing a new standard archive file for crystallography. The two, of course, were not unconnected: the Acta C article was the first to be submitted using the new file format, CIF, described in the Acta A paper. Since that time, CIF has grown to be the accepted standard way to describe crystal and molecular structures derived from single-crystal X-ray diffraction experiments. Many journals will not publish such a structure unless it is accompanied by a CIF; and in many cases, the decision to publish will have included a technical review of the quality of the structure using the IUCr's automated checkCIF procedure.
However, CIF has grown far beyond its original design as a standard file format for single-crystal structure reports, and extensions of the original standard are found in an increasing number of crystallographic and other structural science applications. The acronym now stands for 'Crystallographic Information Framework', in recognition of its application across all of these fields, and many of the Commissions of the IUCr are actively engaged in extending its use within their disparate fields. In an age of data-driven science, crystallography has come to be seen as a pioneer in defining experimental and derived data with the precision and scope necessary to achieve the goals of the FAIR data management strategy - namely, to ensure that data are findable, accessible, interoperable and reusable.
Out of the blue?
As with most great ideas, CIF did not spring into the world fully-formed and with no history (Fig. 1). Of course, crystallography is a discipline blessed by an area of study that - to a first approximation - is inherently orderly and well defined (regular three-dimensional packing of atomic or molecular motifs), and that produces copious results from certain reasonably standard types of experiment (diffraction, whether of X-rays, electrons or neutrons). And classification of crystallographic properties, whether of mineral types, crystal habits, or space group symmetries, has been a key activity for decades, if not centuries.
Even so, efforts towards capturing all the information necessary to repeat a crystal structure determination, especially when electronic computers became a significant research tool, go back further than many people realise. Fig. 1 captures some of the steps in the path towards the creation and early implementation of the CIF standard, based on the current authors' memories. Mario Nardelli, a distinguished and prolific structural chemist, launched the journal Crystal Structure Communications (1972-1983) at his home institution at University of Parma, and developed software (Parma Structural Checking – PARST) to ease the labour of checking the reported structures. When the journal was taken into the IUCr family of publications as Acta Crystallographica Section C, its rigorous standards of checking were adopted by the Editor-in-Chief, Sidney Abrahams, who formalised the requirements for information (what we would now call experimental metadata) needed to check the consistency and reasonableness of the reported structure. Similar requirements were emerging at that time within the relatively new structural databases, such as those developed for inorganic structures (ICSD) and organic/organometallic structures (CSD). Within the Cambridge Crystallographic Data Centre (CCDC), validation software (UNIMOL) was developed and subsequently used for the CSD and in early automated structure checking in IUCr journals.
![[Portrait of Isaac Newton]](https://www.iucr.org/__data/assets/image/0003/152760/isaac-newton-shoulders-of-giants-quote.jpg)
Sir Isaac Newton
Shoulders of Giants
- Mario Nardelli Crystal Structure Communications, PARST
- Sidney Abrahams Acta Crystallographica Section C
- Frank Allen, Peter Murray-Rust, David Watson, Sam Motherwell, Olga Kennard CSD
- I. David Brown, Günter Bergerhoff ICSD
- Jim Stewart XRAY76
- Ted Maslen Working Party on Crystallographic Information
- Syd Hall Xtal, CIF
- Eric Gabe NRCVAX
- George Sheldrick SHELXL
- David Watkin CRYSTALS
- Ton Spek PLATON
- Walter Hamilton, Tom Koetzle, Joel Sussman, Helen Berman PDB, NDB
- Paula Fitzgerald, John Westbrook mmCIF
Special mention should be made of David Brown, of McMaster University, who led the IUCr's first initiative towards a computer-readable standard file format - the Standard Crystallographic File Structure (SCFS)[1]. In the 1980s, when Fortran dominated scientific software development, any such standard was inevitably tied to Fortran input/output conventions (80-column records, fixed field lengths for different types of data); and by the late 1980s this was seen as lacking the flexibility to meet the increasingly complex requirements of increasingly capable and ambitious programming projects. Nevertheless, the SCFS project (informed by the requirements of the databases and journals) identified many of the discrete data items that needed to be transferred between different crystallographic programs, work that was to bear fruit in the development of the CIF core dictionary.
Jim Stewart pioneered a type of structured information storage within the crystallographic software package XRAY that was further refined by Syd Hall (University of Western Australia) in Xtal, an early example of a collaborative project where crystallographic programmers around the world contribute separate modules to a growing library of software. Syd, Frank Allen of the CCDC and David Brown together developed CIF as a standard format in 1991, an outcome of the Working Party on Crystallographic Information that the IUCr had convened in 1987 under the leadership of Ted Maslen. Together with a lightweight extensible free-form structure, CIF was populated by the data items identified as essential to validation and structure characterization in databases and journals. Authors of leading structure refinement software of the time rapidly adopted this standard, giving the impetus for its universal adoption within the small-unit-cell community. Ton Spek developed a very powerful software tool that checked both the internal consistency of the reported structure and its chemical reasonableness, the latter informed by the wealth of data in the structural databases.
![[Portrait montage: David Brown, Syd Hall, Frank Allen]](https://www.iucr.org/__data/assets/image/0008/152783/IDBrown_SRHall_FHAllen.jpg) Figure 2. David Brown, Syd Hall, Frank Allen: authors of the original CIF specification.
Figure 2. David Brown, Syd Hall, Frank Allen: authors of the original CIF specification.
Meanwhile, the Protein Data Bank (PDB), founded in 1971 as a repository for biological macromolecular structures, was also feeling the limitations of the Fortran-style standard that it had adopted. The community embarked upon an extension of CIF – the macromolecular Crystallographic Information File – that could capture all the experimental data associated with a protein structure determination by X-ray diffraction, but that could also adequately describe the complex structure of intricately folded protein and nucleic acid molecules. To meet the needs of the new relational database that was being designed for the PDB, mmCIF imposed more constraints on the relations between the data items that it defined. In a series of workshops[2], the mmCIF standard was refined to become a superset of the core dictionary with the additional attributes that made it formally equivalent to a relational database.
Timeline of Crystallographic Information
This timeline illustrates significant milestones in crystallographic publishing, database development and information management.
CIF and Crystallography
Many young researchers will have grown up with the idea of CIF as the natural way to publish single-crystal structure reports and import structural models; and they will be equally familiar with checkCIF as a validation tool and indicator of the completeness and precision of a structure determination.
While many journals require a CIF as supporting information, Fig. 4 shows the particular power of the complete integration of CIF in the publishing process that IUCr journals offers. In this interactive figure, the author has provided some alternative views of the molecule studied, highlighting different areas of interest. However, the reader can right-click into the main image to find a much larger menu of options, permitting visualization of the unit-cell packing, the crystal structure, or individual symmetry operations; and interrogation of the data for arbitrary geometric measurements.
| |
|
|
|
|
|
However, researchers may have had fewer opportunities to use the small-unit-cell derivatives of CIF that have been developed during their lifetime, and that are gradually becoming established as ways to describe more complex types of structure. There are now separate extension dictionaries that cover such fields as: powder diffraction (embracing the different measurables of a wider range of instrumentation, the practice of using multiple data sets to fit a single crystallographic model, and the need to characterize different phases); modulated and composite structures (with the ability to assign superspace groups and modulation wave vectors); magnetic structures (describing both commensurate and incommensurate magnetic structures exhibiting long-range three-dimensional magnetic order); and the description of the topology of lattices and their relation to crystal structures (Fig. 5).












There are also small extension dictionaries describing aspects of twinning, multipole expansion of electron density, and structure refinement restraints and constraints. These are not very widely used, but are available as starting points for more detailed treatment when required. Many of the IUCr Commissions are also interested in developing CIF dictionaries to provide standard machine representations within their own spheres of interest.
In the area of structural biology, the Worldwide Protein Data Bank curates the mmCIF family of dictionaries, including PDBx (the main extension to mmCIF that tracks the developing field of protein crystallography) and a number of extension dictionaries relevant to NMR structure determinations, small-angle scattering, three-dimensional electron microscopy, integrative/hybrid methods, features of synchrotron radiation facilities and beamlines, and validation reports.
Another important development has been imgCIF, designed to capture diffraction image data and also the necessary experimental metadata allowing proper interpretation of the images captured. Since imgCIF and its binary data representation equivalent CBF (the Crystallographic Binary File) were first developed in the late 1990s, data acquisition volumes and rates have increased so rapidly that this type of file format is no longer suitable for real-time data capture. Nevertheless, the data definitions of imgCIF have partly informed the NeXus macromolecular crystallography application definition (NXmx) that provides full metadata definitions in the HDF5 format increasingly used by beamlines.
This is a significant illustration that the real power of CIF lies in its definitions of concepts and quantities. While the concrete CIF file format is a useful information exchange mechanism, it is relatively easy to translate the format to other common standards, such as XML, the old PDB format, or – increasingly popular – JSON.
A simple catalogue of this plethora of dictionaries and their specific applications perhaps obscures the real importance that CIF has acquired during the course of its evolution. It now touches upon the complete workflow of a structure determination, from the capture of the experimental data, through its interpretation, modelling and publication, to worldwide dissemination in curated databases (Fig. 6). In data science applications, the collection of controlled vocabularies and interrelationships among detailed data definitions has come to be known as an 'ontology', and crystallography now has one of the most completely developed of any science. It is now possible to offer schools and workshops to early-career structural scientists where experimental best practice is developed with the full rigour of data characterization, interpretation and validation[5].
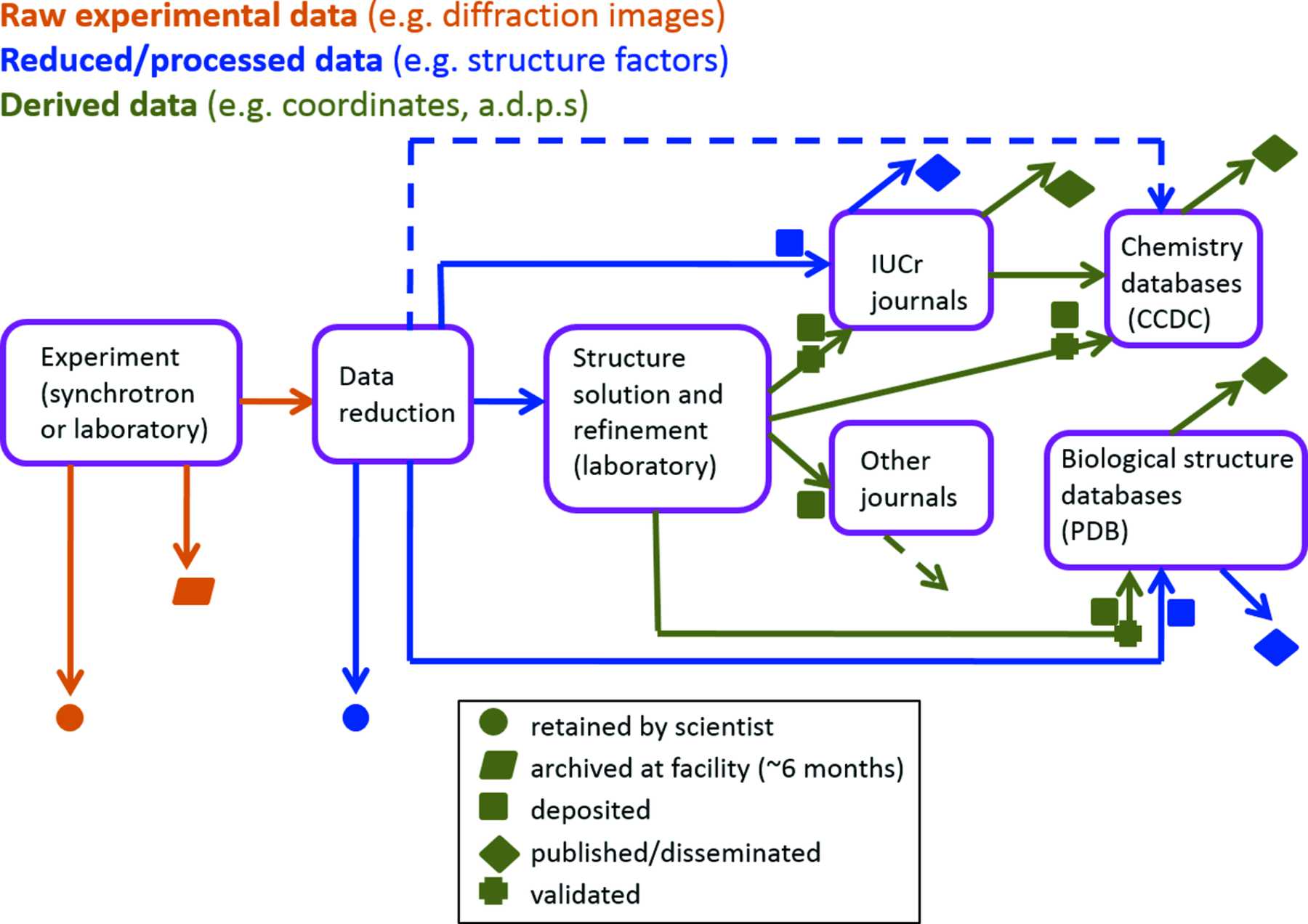 Figure 6. A coherent information flow in crystallography. CIF ontologies characterize data at every stage of the information processing life cycle, from experimental apparatus to published paper and curated database deposit.
Figure 6. A coherent information flow in crystallography. CIF ontologies characterize data at every stage of the information processing life cycle, from experimental apparatus to published paper and curated database deposit.
Leading the way
The significance of CIF has been recognised in the information and data science communities, in the form of the 2006 Association of Learned and Professional Society Publishers Award for Publishing Innovation and the prestigious 2014 CODATA Prize to Professor Sydney Hall. In both cases the award citations were generous in their acclaim for what the CODATA judges called 'a momentous contribution'.
Yet despite the potential applicability of the approach to any field of science, there has been relatively little penetration in other scientific domains. Syd Hall intended CIF to be just one application of a general approach – 'STAR' (Self-Defining Text Archive and Retrieval) – which has also been used in small-scale pilots in chemistry (MIF, the molecular information file), quantum chemistry, and botany, and most successfully as the basis for the NMR structures database of BioMagResBank, one of the partners of the wwPDB[6].
There are also encouraging signs of novel projects in solid-state science that are inspired by CIF[7]. These are greatly to be welcomed, as a proliferation of domain ontologies in a common format will certainly lead to easier interoperability. It is likely that, despite its success within crystallography, many other disciplines consider the STAR/CIF approach as niche, and not sufficiently supported by the wider information and data science communities. Nevertheless, in our opinion as people who have been involved with CIF for a combined 45 years, STAR and CIF have much to offer, not least in the process of devising new ontologies. The basic file syntax is lightweight and clean, and the dictionary attribute sets (that is, the terms used to express definitions of concepts in machine-readable ways) are not unduly complex, and extensible where needed.
Relationships may be expressed in many different ways – early interest in the STAR File with its nested loops that are absent from CIF explored its suitability for populating object-relational databases. Yet the more natural relational-database type structure that CIF more easily encapsulates is adequate for developing conceptual frameworks of sufficient complexity for most scientific purposes[8]. We respectfully encourage scientists who need to provide machine-readable descriptions of scientific data and metadata within their own discipline to consider the STAR/CIF approach as a useful starting point. Development into more complex web ontology frameworks such as OWL[9], if needed, can be left to a later implementation stage, once the essential definitions and relationships have been expressed in the manner of a CIF dictionary.
Constancy and change
As is apparent from the title of the 1991 CIF paper, one of its primary design goals was to archive data. As such, stability is crucial, both of the file format and of the dictionary definitions. Definitions should not be changed, as that could invalidate archived data sets. On the other hand, concepts do evolve, and the dictionaries will acquire new data names with their associated definitions. Where there is a clear shift in the way an existing concept is realised, there are mechanisms to mark an old data name as deprecated and to express its relationship to a new entry.
In fact, the core dictionary has grown rather little since its original release, reflecting the relative maturity of single-crystal structure determination. In marked contrast, the PDBx extension to the original mmCIF dictionary has raised the number of data names defined for the core biological macromolecular structure determination from around 1670 to over 6400, reflecting both the complexity of the structural description of proteins and nucleic acids and the explosive growth of the subject itself.
To accommodate new requirements in data management the CIF file format itself was revised in 2016[10]. Its main differences from version 1 were the adoption of Unicode as the native character set, the ability to represent all possible text strings, and simpler ways to represent vectors, matrices and other compound data structures. This was coincident with the adoption of a new formalism for dictionary definitions known as DDLm, a methods-capable dictionary definition language. The term 'methods' indicates that formal relationships between different data items can be expressed, evaluated and validated by machine-executable statements within the dictionary itself. This opens the way to more rigorous validation tools, and in principle brings closer the idea of a universal processing engine that can manipulate scientific entities by inputing a suitable dictionary – no domain-specific coding would be needed within such programs.
Again reflecting the maturity of single-crystal diffraction and the existence of established software that adequately handles the publication requirements of such structures, relatively few 'CIF2' files are yet found in the wild. However, we expect that they will be popular in newer areas of research, and we note, for example, the adoption of CIF2 in a novel Raman spectroscopy database[11].
The image we have chosen to introduce this article is, we feel, an appropriate metaphor for CIF at the end of its first three decades of deployment. From small beginnings, it has grown to a significant size, constantly propelled by a small core of developers and adopters. In the process, it may have acquired some grit and irregularities around the edge, and is now at a stage where even more effort will be needed, especially to gather up more material from the fresh 'snowfields' of new techniques, structural representations, and understanding.
However, most of the pioneers from the early days have now retired or otherwise withdrawn from the scene. If we are not to lose momentum, we need fresh young blood to keep the snowball rolling and growing ever larger. Please join in the fun!
Resources

Find out more about CIF:
- The IUCr CIF website
- PDBX/mmCIF dictionary resources maintained by wwPDB
- Crystallographic Information and Data Management Symposium, U. Warwick, 2013
- Crystallographic Information Fiesta School, Naples 2019
- International Tables for Crystallography Volume G: Definition and exchange of crystallographic data (1st online edition 2006; a second edition is in preparation)
Notes and references
[1] Brown, I. D. (1988). Standard Crystallographic File Structure-87. Acta Cryst. A44, 232. DOI: https://doi.org/10.1107/S010876738700970X
[2] Fitzgerald, P., Berman, H., Bourne, P., McMahon, B., Watenpaugh, K. & Westbrook, J. (1996). The macromolecular CIF dictionary https://www.iucr.org/resources/commissions/crystallographic-computing/schools/school96/the-mmcif-dictionary
[3] Knott, S. A., Hitchcock, S. R. & Ferrence, G. M. (2008). (5S,6S)-4,5-Dimethyl-3-methylacryloyl-6-phenyl-1,3,4-oxadiazinan-2-one. Acta Cryst. E64, o1101. DOI: https://doi.org/10.1107/S1600536808013986
[4] Jsmol is the HTML5 modality of Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org/ Under continual development by its principal developer, Robert M. Hanson, this tool provides innovative and insightful visualizations of structures described by many flavours of CIF, including macromolecular (mmCIF), incommensurate modulated (msCIF), magnetic (magCIF) and topological (topoCIF) structures.
[5] The pioneering CIFiesta school, hosted by the Italian Crystallographic Association in Naples in 2019, has been described in the IUCr Newsletter. Teaching materials from the school are available at: https://www.iucr.org/resources/cif/comcifs/cifiesta-2019
[6] Hall, S.R. & McMahon, B. (2016). The Implementation and Evolution of STAR/CIF Ontologies: Interoperability and Preservation of Structured Data. Data Science Journal, 15, p.3.
[7] Some examples of recent standards development efforts in solid-state science: Zainul Ihsan, A., Dessi, D., Alam, M., Sack, H. & Sandfeld, S. (2021). Steps towards a Dislocation Ontology for Crystalline Materials. arXiv [cond-mat.mtrl-sci], arXiv:2106.15136; Evans, J. D., Bon, V., Senkovska, I. & Kaskel, S. (2021). A Universal Standard Archive File for Adsorption Data. Langmuir, 37, 4222–4226. DOI: https://doi.org/10.1021/acs.langmuir.1c00122; Andersen, C. W., Armiento, R., Blokhin, E. et al. (2021). OPTIMADE, an API for exchanging materials data. Sci Data 8, 217. DOI: https://doi.org/10.1038/s41597-021-00974-z; Cheung, K., Drennan, J. & Hunter, J. (2008). Towards an ontology for data-driven discovery of new materials. AAAI Spring Symposium: Semantic Scientific Knowledge Integration, pp. 9–14.
[8] Hester, J. (2016). A Robust, Format-Agnostic Scientific Data Transfer Framework. Data Science Journal, 15, p.12. DOI: http://doi.org/10.5334/dsj-2016-012
[9] Hitzler, P., Krötzsch, M., Parsia, B., Patel-Schneider, P. & Rudolph, S. (2012). OWL 2 Web Ontology Language Primer In: Tech. rep. W3C. Available at: https://www.w3.org/TR/2012/REC-owl2-primer-20121211.
[10] Bernstein, H. J., Bollinger, J. C., Brown, I. D., Grazulis, S., Hester, J. R., McMahon, B., Spadaccini, N., Westbrook, J. D. & Westrip, S. P. (2016). Specification of the Crystallographic Information File format, version 2.0. J. Appl. Cryst. 49, 277–284. DOI: https://doi.org/10.1107/S1600576715021871
[11] El Mendili, Y., Vaitkus, A., Merkys, A., Grazulis, S., Chateigner, D., Mathevet, F., Gascoin, S., Petit, S., Bardeau, J.-F., Zanatta, M., Secchi, M., Mariotto, G., Kumar, A., Cassetta, M., Lutterotti, L., Borovin, E., Orberger, B., Simon, P., Hehlen, B. & Le Guen, M. (2019). Raman Open Database: first interconnected Raman–X-ray diffraction open-access resource for material identification. J. Appl. Cryst. 52, 618–625. DOI: https://doi.org/10.1107/S1600576719004229
Appreciation
As we were working on this article, we were saddened to hear of the unexpected passing of John Westbrook (1957–2021), who did so much to develop the mmCIF/PDBx and related extension standards, and whose companionship along almost the entire route of CIF development will be greatly missed.
Brian McMahon is based at the IUCr offices in Chester, UK, and has been COMCIFS Secretary since 1993. James Hester works at ANSTO, Australia, and has been COMCIFS Chair since 2008.
Copyright © - All Rights Reserved - International Union of Crystallography








